
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,894,978 – 26,895,083 |
| Length | 105 |
| Max. P | 0.579119 |

| Location | 26,894,978 – 26,895,083 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 84.36 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -27.24 |
| Energy contribution | -26.84 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
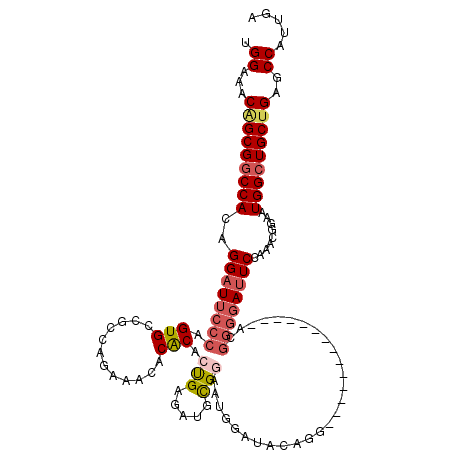
>3R_DroMel_CAF1 26894978 105 + 27905053 UCAAUGGCUCAGCAGCCAUUCCGUUUGGAAUCCCCGU--------------CCUGUAUCCAUUUCGCAUCUCAGUGUGUGCUUCUGGCGGCACUGGGAAUCCUGUGGCCGCUGUUUCCA ....(((..((((.(((((.......(((.((((.((--------------.((((........(((((....)))))........)))).)).)))).))).))))).))))...))) ( -36.19) >DroSec_CAF1 3775 105 + 1 UCAAUGGCUCAGCAGCCAUUCCGUUUGGAAUCCCCGU--------------CCUGUAUCCAUUCCGCAUCUCGGUGCGUUUUUCUGGCGGCACUGGGAAUCCUGUGGCCGCUGUUUCCA ....(((..((((.(((((.......(((.((((.((--------------.((((........(((((....)))))........)))).)).)))).))).))))).))))...))) ( -38.89) >DroSim_CAF1 4579 105 + 1 UCAAUGGCUCAGCAGCCAUUCCGUUUGGAAUCCCCGU--------------CCUGUAUCCAUUCCGCAUCUCGGUGCGUGUUUCUGGCGGCACUGGGAAUCCUGUGGCCGCUGUUUCCA ....(((..((((.(((((.......(((.((((.((--------------.((((........(((((....)))))........)))).)).)))).))).))))).))))...))) ( -38.89) >DroEre_CAF1 3846 96 + 1 UCAAUGGCUCAGCAGCCAUUCCGCUUCGAGU--------------------CCUGUAUUCAUUUCGCAUUUCAGUGUGCA---CUGGCGGCACUGGGAAUCCUGUGGUCGCAGUUUCCG ..(((((((....)))))))(((((..((((--------------------.....)))).....((((......)))).---..))))).....((((..((((....)))).)))). ( -27.40) >DroYak_CAF1 3996 119 + 1 UCAAUGGCUCAGCAGCCAUUCCGUUUCGAAUCCCCGUUCCCGUCCCCGUAUCCUGCAUCCUUUCCACAUCUCAGUGUGUGUUUCUGGCGGCACUGGGAAUCCUGUGGCCGCCGUUUCCA ..((((((...((((............((((....)))).............)))).......((((((((((((((.(((.....))))))))))))....)))))..)))))).... ( -30.11) >consensus UCAAUGGCUCAGCAGCCAUUCCGUUUGGAAUCCCCGU______________CCUGUAUCCAUUCCGCAUCUCAGUGUGUGUUUCUGGCGGCACUGGGAAUCCUGUGGCCGCUGUUUCCA ..(((((((....))))))).....(((((..................................(((((....))))).......((((((((.((....)).)).))))))..))))) (-27.24 = -26.84 + -0.40)



| Location | 26,894,978 – 26,895,083 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 84.36 |
| Mean single sequence MFE | -36.46 |
| Consensus MFE | -22.78 |
| Energy contribution | -23.82 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26894978 105 - 27905053 UGGAAACAGCGGCCACAGGAUUCCCAGUGCCGCCAGAAGCACACACUGAGAUGCGAAAUGGAUACAGG--------------ACGGGGAUUCCAAACGGAAUGGCUGCUGAGCCAUUGA (((...(((((((((......((((.((.((.(((...((((.......).)))....))).....))--------------)).))))((((....)))))))))))))..))).... ( -37.70) >DroSec_CAF1 3775 105 - 1 UGGAAACAGCGGCCACAGGAUUCCCAGUGCCGCCAGAAAAACGCACCGAGAUGCGGAAUGGAUACAGG--------------ACGGGGAUUCCAAACGGAAUGGCUGCUGAGCCAUUGA (((...(((((((((......((((.((.((.(((......((((......))))...))).....))--------------)).))))((((....)))))))))))))..))).... ( -39.60) >DroSim_CAF1 4579 105 - 1 UGGAAACAGCGGCCACAGGAUUCCCAGUGCCGCCAGAAACACGCACCGAGAUGCGGAAUGGAUACAGG--------------ACGGGGAUUCCAAACGGAAUGGCUGCUGAGCCAUUGA (((...(((((((((......((((.((.((.(((......((((......))))...))).....))--------------)).))))((((....)))))))))))))..))).... ( -39.60) >DroEre_CAF1 3846 96 - 1 CGGAAACUGCGACCACAGGAUUCCCAGUGCCGCCAG---UGCACACUGAAAUGCGAAAUGAAUACAGG--------------------ACUCGAAGCGGAAUGGCUGCUGAGCCAUUGA .((((.(((......)))..)))).....(((((((---(....)))).....(((..((....))..--------------------..)))..))))(((((((....))))))).. ( -25.00) >DroYak_CAF1 3996 119 - 1 UGGAAACGGCGGCCACAGGAUUCCCAGUGCCGCCAGAAACACACACUGAGAUGUGGAAAGGAUGCAGGAUACGGGGACGGGAACGGGGAUUCGAAACGGAAUGGCUGCUGAGCCAUUGA (((...(((((((((..((((((((..(((..((.....((((........))))....))..))).....((....)).....)))))))).........)))))))))..))).... ( -40.40) >consensus UGGAAACAGCGGCCACAGGAUUCCCAGUGCCGCCAGAAACACACACUGAGAUGCGGAAUGGAUACAGG______________ACGGGGAUUCCAAACGGAAUGGCUGCUGAGCCAUUGA .((...(((((((((..((((((((.(((............))).(((.....)))............................)))))))).........)))))))))..))..... (-22.78 = -23.82 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:47:34 2006