| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,887,188 – 26,887,304 |
| Length | 116 |
| Max. P | 0.901685 |

| Location | 26,887,188 – 26,887,304 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 65.04 |
| Mean single sequence MFE | -36.44 |
| Consensus MFE | -15.38 |
| Energy contribution | -15.58 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.42 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.854304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
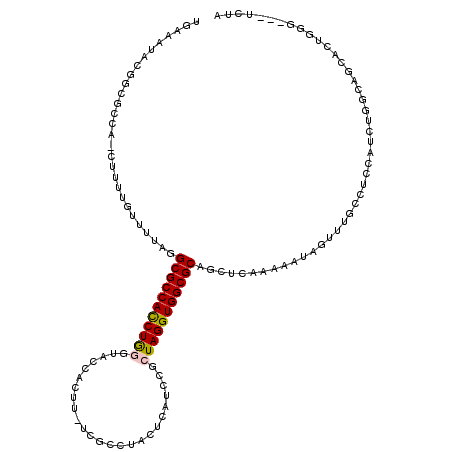
>3R_DroMel_CAF1 26887188 116 + 27905053 UAGA---CCCAGUGCUGCCAGAUGGACGAAAAAUAUUUUUGAGCUGCGCCACCUAGCGGAUGAGUUGGCGA-AAGUGGUAUCCAGGUGGCGCCCGAAACAAAAGGUGGCGCCUUAUUUCA ....---....((((..((.......(((((.....)))))....(((((((((...(((((...(.((..-..)).)))))))))))))))...........))..))))......... ( -39.50) >DroSec_CAF1 95449 108 + 1 UACA---CCCAGUGCUGCCAGAUGGAGGCAAACUACUUGUGAGCUGCGCCACCUAGCGGAUGAGUACGCGAAAAGUGGUACCCAGAUGGCGCCUAAUAUU---------UCUGUAUUUCU ....---...((.((.((((..(((.(((..((.....))..)))..(((((...(((........))).....)))))..)))..))))))))......---------........... ( -29.40) >DroSim_CAF1 99139 108 + 1 CAGA---CCCAGUGCUGCCAGAUGGAGGCAAACUACUUGUAAGCUGCGCCACCUAGCGGAUGAGUAGGCGAAAAGUGGUACCCAGGUGGCGCCUACAAUU---------UCAGUAUAUCU ....---....(((((((.((.(((.(((..((.....))..)))...))).)).))(((...(((((((..(..(((...)))..)..)))))))...)---------))))))).... ( -31.40) >DroEre_CAF1 104142 119 + 1 UAACCUGGCCAGUGUUGCCAGAUAGGCGCAAGCUAUUUUGGAAGCGCGCCACCUAGCGGUCGUACAUGCGA-AUGUGACAUAUAGGUGGCGCUUCAAGCAAAAGUUGGCGCAAUAUUUCA .....(((((((.(((.(((((..(((....)))..))))).)))((((((((((...(((..((((....-)))))))...))))))))))............))))).))........ ( -47.50) >DroYak_CAF1 110411 92 + 1 UAA---------------------------AAAUAUUUUUAAGGUGCGCCACCUAGCGGAUGCCCAGACGA-AUGUGGUACAUAGGUGGCGCACAAAACAAAAGGUGGCGCCAUUUUUCA ...---------------------------.............((((((((((((...(.((((((.....-.)).))))).))))))))))))......(((((((....))))))).. ( -34.40) >consensus UAAA___CCCAGUGCUGCCAGAUGGACGCAAACUAUUUUUGAGCUGCGCCACCUAGCGGAUGAGUAGGCGA_AAGUGGUACCCAGGUGGCGCCUAAAACAAAAG_UGGCGCAAUAUUUCA .............................................(((((((((.((......))..((.....)).......)))))))))............................ (-15.38 = -15.58 + 0.20)


| Location | 26,887,188 – 26,887,304 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 65.04 |
| Mean single sequence MFE | -33.65 |
| Consensus MFE | -19.13 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26887188 116 - 27905053 UGAAAUAAGGCGCCACCUUUUGUUUCGGGCGCCACCUGGAUACCACUU-UCGCCAACUCAUCCGCUAGGUGGCGCAGCUCAAAAAUAUUUUUCGUCCAUCUGGCAGCACUGGG---UCUA ........((((((.............))))))((((((((.......-..........))))(((((((((((..................)).)))))))))......)))---)... ( -31.62) >DroSec_CAF1 95449 108 - 1 AGAAAUACAGA---------AAUAUUAGGCGCCAUCUGGGUACCACUUUUCGCGUACUCAUCCGCUAGGUGGCGCAGCUCACAAGUAGUUUGCCUCCAUCUGGCAGCACUGGG---UGUA ...........---------........((((((((((((((((.......).)))).......))))))))))).....(((..(((((((((.......))))).))))..---))). ( -32.91) >DroSim_CAF1 99139 108 - 1 AGAUAUACUGA---------AAUUGUAGGCGCCACCUGGGUACCACUUUUCGCCUACUCAUCCGCUAGGUGGCGCAGCUUACAAGUAGUUUGCCUCCAUCUGGCAGCACUGGG---UCUG ((((.......---------..(((((((((((((((((.........................)))))))))...)))))))).(((((((((.......))))).)))).)---))). ( -34.31) >DroEre_CAF1 104142 119 - 1 UGAAAUAUUGCGCCAACUUUUGCUUGAAGCGCCACCUAUAUGUCACAU-UCGCAUGUACGACCGCUAGGUGGCGCGCUUCCAAAAUAGCUUGCGCCUAUCUGGCAACACUGGCCAGGUUA .........((((...............((((((((((...(((((((-....))))..)))...))))))))))(((........)))..))))..(((((((.......))))))).. ( -41.40) >DroYak_CAF1 110411 92 - 1 UGAAAAAUGGCGCCACCUUUUGUUUUGUGCGCCACCUAUGUACCACAU-UCGUCUGGGCAUCCGCUAGGUGGCGCACCUUAAAAAUAUUU---------------------------UUA .........(((((((((...((...((((.(((...(((.....)))-.....)))))))..)).)))))))))...............---------------------------... ( -28.00) >consensus UGAAAUACGGCGCCA_CUUUUGUUUUAGGCGCCACCUGGGUACCACUU_UCGCCUACUCAUCCGCUAGGUGGCGCAGCUCAAAAAUAGUUUGCCUCCAUCUGGCAGCACUGGG___UCUA ............................(((((((((((.........................)))))))))))............................................. (-19.13 = -19.13 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:47:29 2006