
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,814,492 – 26,814,583 |
| Length | 91 |
| Max. P | 0.768864 |

| Location | 26,814,492 – 26,814,583 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -24.07 |
| Consensus MFE | -10.57 |
| Energy contribution | -10.77 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.768864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
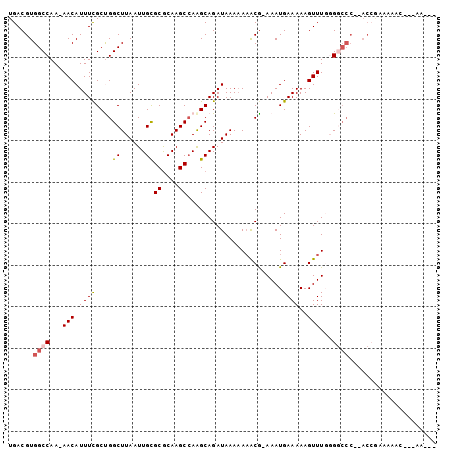
>3R_DroMel_CAF1 26814492 91 + 27905053 UGACGUGGCCAAAAACAUUUCGCUGGCUUAAUUGCGCGCAAGCCAAGCAGAUAAAAAAACA-AAAUGAAAAAGUUUGGGGCCC--ACCGAAAAA--------- ....((((((..((((.....((((((((..........))))).))).............-...(....).))))..)).))--)).......--------- ( -20.10) >DroPse_CAF1 54978 98 + 1 UGACGUGGCCAA-AACAUUUUGCCGGCUUAAUUGUGCGCAAGCCAAGCAGAUAAA-AAACGAAAAUGAAAAAGUUUUCGGCCCCAACCGCCAACCGUCAA--- ((((((((((((-(....))))..(((....(((.((....))))).........-...(((((((......))))))))))......))))..))))).--- ( -27.30) >DroSec_CAF1 23142 78 + 1 UGACGUGGCCAAAAACAUUUCGCUGGCUUAAUUGCGCGCAAGCCAAGCAGAUAAAAAAGCG-AAAUGAAAAAGUUUGGG------------------------ ........(((((..(((((((((...(((.((((((....))...)))).)))...))))-)))))......))))).------------------------ ( -24.20) >DroEre_CAF1 27467 97 + 1 UGACGUGGCCAAAAACAUUUCGCUGGCUUAAUUGCGCGCAAGCAAAGCAGAUAAAAAAGCG-AAAUGAAAAAGUUUGGGGCCC--ACCAAAAAACAAAAA--- ....(((((((((..(((((((((...(((.((((((....))...)))).)))...))))-)))))......)))))...))--)).............--- ( -28.20) >DroAna_CAF1 27121 98 + 1 UGACGUGGCCAA-AACAUUUCGCUGGCUUAAUUGUGCACAAGCCAGGCAGAUAAAAA-ACG-AAAUGAAAAAGUUUGGGUCCC--ACAAAAAAGUUGAAAAAA ....((((((((-(.((((((((((((((..........)))))))...........-.))-)))))......)))))...))--))................ ( -21.70) >DroPer_CAF1 55341 99 + 1 UGACGUGGCCAA-AACAUUUUGCCGGCUUAAUUGUGCGCAAGCCAAGCAGAUAAAAAAACGAAAAUGAAAAAGUUUUCGGCCCCAACCGCCAACCGCCAA--- .....(((((((-(....))))..(((....(((.((....))))).............(((((((......))))))).........)))....)))).--- ( -22.90) >consensus UGACGUGGCCAA_AACAUUUCGCUGGCUUAAUUGCGCGCAAGCCAAGCAGAUAAAAAAACG_AAAUGAAAAAGUUUGGGGCCC__ACCGAAAAAC___AA___ ......((((...(((.((((....((......))((....)).......................))))..)))...))))..................... (-10.57 = -10.77 + 0.19)


| Location | 26,814,492 – 26,814,583 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.38 |
| Mean single sequence MFE | -24.53 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.45 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
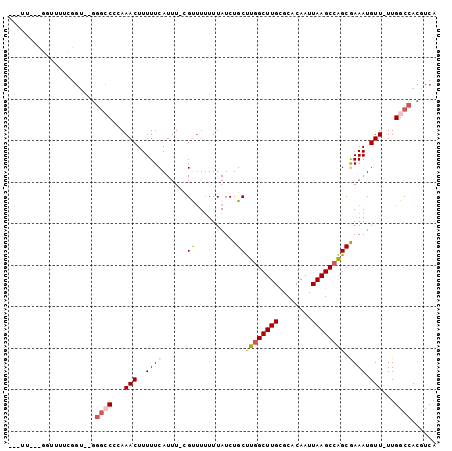
>3R_DroMel_CAF1 26814492 91 - 27905053 ---------UUUUUCGGU--GGGCCCCAAACUUUUUCAUUU-UGUUUUUUUAUCUGCUUGGCUUGCGCGCAAUUAAGCCAGCGAAAUGUUUUUGGCCACGUCA ---------......(..--(((((..((((..((((....-((......))...(((.((((((........))))))))))))).))))..)))).)..). ( -23.20) >DroPse_CAF1 54978 98 - 1 ---UUGACGGUUGGCGGUUGGGGCCGAAAACUUUUUCAUUUUCGUUU-UUUAUCUGCUUGGCUUGCGCACAAUUAAGCCGGCAAAAUGUU-UUGGCCACGUCA ---.((((((....)(((..((((((((((........))))))...-......((((.((((((........))))))))))....)))-)..))).))))) ( -28.50) >DroSec_CAF1 23142 78 - 1 ------------------------CCCAAACUUUUUCAUUU-CGCUUUUUUAUCUGCUUGGCUUGCGCGCAAUUAAGCCAGCGAAAUGUUUUUGGCCACGUCA ------------------------.(((((......(((((-((((..((((..(((.((.....)).)))..))))..)))))))))..)))))........ ( -19.30) >DroEre_CAF1 27467 97 - 1 ---UUUUUGUUUUUUGGU--GGGCCCCAAACUUUUUCAUUU-CGCUUUUUUAUCUGCUUUGCUUGCGCGCAAUUAAGCCAGCGAAAUGUUUUUGGCCACGUCA ---.............((--((...(((((......(((((-((((..((((..(((...((....)))))..))))..)))))))))..))))))))).... ( -24.60) >DroAna_CAF1 27121 98 - 1 UUUUUUCAACUUUUUUGU--GGGACCCAAACUUUUUCAUUU-CGU-UUUUUAUCUGCCUGGCUUGUGCACAAUUAAGCCAGCGAAAUGUU-UUGGCCACGUCA ................((--((...(((((......(((((-((.-...........((((((((........))))))))))))))).)-)))))))).... ( -23.70) >DroPer_CAF1 55341 99 - 1 ---UUGGCGGUUGGCGGUUGGGGCCGAAAACUUUUUCAUUUUCGUUUUUUUAUCUGCUUGGCUUGCGCACAAUUAAGCCGGCAAAAUGUU-UUGGCCACGUCA ---.((((((....)(((..((((((((((........))))))..........((((.((((((........))))))))))....)))-)..))).))))) ( -27.90) >consensus ___UU___GGUUUUCGGU__GGGCCCCAAACUUUUUCAUUU_CGUUUUUUUAUCUGCUUGGCUUGCGCACAAUUAAGCCAGCGAAAUGUU_UUGGCCACGUCA .....................((((...(((..((((......((..........))((((((((........)))))))).)))).)))...))))...... (-12.86 = -13.45 + 0.59)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:47:13 2006