
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,780,368 – 26,780,506 |
| Length | 138 |
| Max. P | 0.996118 |

| Location | 26,780,368 – 26,780,474 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -39.06 |
| Consensus MFE | -24.09 |
| Energy contribution | -24.17 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
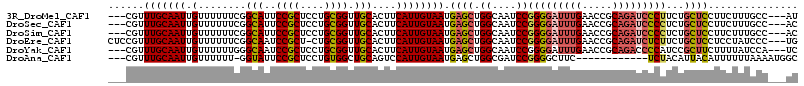
>3R_DroMel_CAF1 26780368 106 + 27905053 GCGGU-----------UUGUGGUGUGGUGGGUGGUAUGGGAUGGCAAAGAAGGAGCAGAAGGGAUCUGCGGUUCAAAUCCCCGGAUUGCCAGCUCAUUACAAUGAAGUGCAACCGCA (((((-----------(.((..((((((((((((((.(((((......(((...(((((.....)))))..)))..))))).....))))).))))))))).......)))))))). ( -38.00) >DroSec_CAF1 83701 92 + 1 GCGGC-----------UUGUGGUGG--------------GGUGGCAAAGAAGGAGCAGAGGGGAUCUGCGGUUCAAAUCCCCGGAUUGCCAGCUCAUUACAAUGAAGUGCAACCGCA ((((.-----------(((((((((--------------(.((((((....((.(((((.....)))))((.......))))...)))))).))))))))))((.....)).)))). ( -37.50) >DroSim_CAF1 87483 102 + 1 GCGGC-----------UUGUGGUGGGGU----GUGAUGGGGUGGCAAAGAAGGAGCAGAGGGGAUCUGCGGUUCAAAUCCCCGGAUUGCCAGCUCAUUACAAUGAAGUGCAACCGCA ((((.-----------(((((.(....(----((((((((.((((((....((.(((((.....)))))((.......))))...)))))).)))))))))....).))))))))). ( -44.70) >DroEre_CAF1 85268 92 + 1 GCGGC-----------UUGUGGUGG--------------GCAGGGAUAGGAGGAGCAGAAGAGAUCUGCGGUUCAAAUCCCCGGAUUGCCAGCUCAUUACAAUGAAGUGCAACCGCA ((((.-----------(((((((((--------------((.(((((..(((..(((((.....)))))..)))..))))).((....)).)))))))))))((.....)).)))). ( -38.70) >DroYak_CAF1 87016 103 + 1 GCGGUGGGCAUGGAUAUGGAGAUGG--------------GGAUGGAUAAAAGAAGCGGAUGGGGUCUGCGGUUCAAAUCCCCGGAUUGCCAGCUCAUUACAAUGAAGUGCAACCGCA (((((..((((.....(((...(((--------------((((((((.......((((((...)))))).)))))..)))))).....)))..((((....)))).)))).))))). ( -36.40) >consensus GCGGC___________UUGUGGUGG______________GGUGGCAAAGAAGGAGCAGAAGGGAUCUGCGGUUCAAAUCCCCGGAUUGCCAGCUCAUUACAAUGAAGUGCAACCGCA ((((............((((((((..............................(((((.....)))))(((...((((....)))))))....))))))))..........)))). (-24.09 = -24.17 + 0.08)


| Location | 26,780,368 – 26,780,474 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -23.72 |
| Energy contribution | -23.40 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.985070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26780368 106 - 27905053 UGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCCCUUCUGCUCCUUCUUUGCCAUCCCAUACCACCCACCACACCACAA-----------ACCGC .(((((((((......))).(((.(.((((((...(((((........(((((.....))))))))))..)))))).).)))..................)-----------))))) ( -27.60) >DroSec_CAF1 83701 92 - 1 UGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCCCCUCUGCUCCUUCUUUGCCACC--------------CCACCACAA-----------GCCGC .((((((..........((((.(((..((((....(((((((((.....)))))))))..))))..))).))))....--------------........)-----------))))) ( -29.35) >DroSim_CAF1 87483 102 - 1 UGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCCCCUCUGCUCCUUCUUUGCCACCCCAUCAC----ACCCCACCACAA-----------GCCGC .((((((.........(((.(((.(.((((((...(((((((((.....)))))))))............)))))).).))).))----)..........)-----------))))) ( -31.57) >DroEre_CAF1 85268 92 - 1 UGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCUCUUCUGCUCCUCCUAUCCCUGC--------------CCACCACAA-----------GCCGC .((((((((..(((((....)))))...))....(((((((..((...(((((.....)))))...))..))))))).--------------........)-----------))))) ( -30.50) >DroYak_CAF1 87016 103 - 1 UGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGACCCCAUCCGCUUCUUUUAUCCAUCC--------------CCAUCUCCAUAUCCAUGCCCACCGC .((((((((..(((((....)))))...)))....((((((.((((..((.((.....)).))....))))...))))--------------))..................))))) ( -23.00) >consensus UGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCCCCUCUGCUCCUUCUUUGCCACC______________CCACCACAA___________GCCGC .(((((((((......))))..((((.((....))(((((((((.....)))))))))...))))...............................................))))) (-23.72 = -23.40 + -0.32)



| Location | 26,780,397 – 26,780,506 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.24 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26780397 109 - 27905053 ---CGUUUGCAAUUGUUUUUUCGGCAUUCCGCUCCUGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCCCUUCUGCUCCUUCUUUGCC---AU ---....(((((((((.....(((....))).....))))))))).......((((.(((..((((....(((((((((.....)))))))))..))))..))).)))).---.. ( -32.60) >DroSec_CAF1 83716 109 - 1 ---CGUUUGCAAUUGUUUUUUCGGCAUUCCGCUCCUGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCCCCUCUGCUCCUUCUUUGCC---AC ---....(((((((((.....(((....))).....))))))))).......((((.(((..((((....(((((((((.....)))))))))..))))..))).)))).---.. ( -34.50) >DroSim_CAF1 87508 109 - 1 ---CGUUUGCAAUUGUUUUUUCGGCAUUCCGCUCCUGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCCCCUCUGCUCCUUCUUUGCC---AC ---....(((((((((.....(((....))).....))))))))).......((((.(((..((((....(((((((((.....)))))))))..))))..))).)))).---.. ( -34.50) >DroEre_CAF1 85283 111 - 1 CUCCGUUUGCAAUUGUUUUUUCGGCAAUCCGCU-CUGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCUCUUCUGCUCCUCCUAUCCC---UG ....((..((.............((((.((((.-..))))))))...((((....))))))..))....(((((((..((...(((((.....)))))...))..)))))---)) ( -33.60) >DroYak_CAF1 87042 109 - 1 ---CGUUUGCAAUUGUUUUUUGGGCAAUCCGCUCCUGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGACCCCAUCCGCUUCUUUUAUCCA---UC ---...(((((((.(......((((((.((((....)))))))).)).)))))))).((((.((....))((((.((((.....))))))))....))))..........---.. ( -28.40) >DroAna_CAF1 98057 99 - 1 ---CGUUUGCAAUUGUUUUUU-GGUAUUCCGCUCCUGUGGCUGCAGUCCAUUGUAAUGAGCUGGCGAUCCGGGGCUUC------------UCUACAUUACAUUUUUUAAAAUGGC ---(((((.....(((..(.(-((......((.((((.(..(((((..(((....)))..)).)))..)))))))...------------.))).)..))).......))))).. ( -19.80) >consensus ___CGUUUGCAAUUGUUUUUUCGGCAUUCCGCUCCUGCGGUUGCACUUCAUUGUAAUGAGCUGGCAAUCCGGGGAUUUGAACCGCAGAUCCCUUCUGCUCCUUCUUUGCC___AC ......(((((((.(........(((..((((....)))).)))....)))))))).((((.((....))(((((((((.....)))))))))...))))............... (-23.20 = -23.80 + 0.60)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:54 2006