
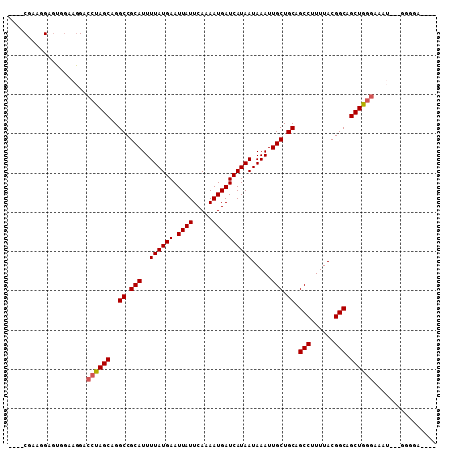
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,774,374 – 26,774,475 |
| Length | 101 |
| Max. P | 0.828342 |

| Location | 26,774,374 – 26,774,475 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -26.40 |
| Consensus MFE | -24.04 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.828342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26774374 101 + 27905053 ----CGAAGGAGUGGAAGGACCCAGCAGGCCGCAUUUUAUGAAUUAUUCAAAAUGAUCAUAAUAAAUUGCUGCAGCCUUUUACGGCAGCUGGGAAAAAGGGGGGA---- ----................((((((..((.(((..((((((.((((.....)))))))))).....))).)).(((......))).))))))............---- ( -27.40) >DroSec_CAF1 77774 98 + 1 ----CGAAGGAGUGGAAGGACCUAGCAGGCCGCAUUUUAUGAAUUAUUCAAAAUGAUCAUAAUAAAUUGCUGCAGCCUUUUACGGCAGCUGGGAAAU---GGGGA---- ----................((((((..((.(((..((((((.((((.....)))))))))).....))).)).(((......))).))))))....---.....---- ( -25.10) >DroSim_CAF1 81483 98 + 1 ----CGAAGGAGUGGAAGGACCUAGCAGGCCGCAUUUUAUGAAUUAUUCAAAAUGAUCAUAAUAAAUUGCUGCAGCCUUUUACGGCAGCUGGGAAAU---GGGGA---- ----................((((((..((.(((..((((((.((((.....)))))))))).....))).)).(((......))).))))))....---.....---- ( -25.10) >DroEre_CAF1 79301 98 + 1 ----CGAAGGAGUAGAAGGACCUAGCAGGCCGCAUUUUAUGAAUUAUUCAAAAUGAUCAUAAUAAAUUGCUGCAGCCUUUUACGGCAGCUGCAAAAU---GGGGC---- ----...(((..........))).....(((.((((((((((.((((.....))))))))))....((((.((.(((......))).)).)))))))---).)))---- ( -23.40) >DroYak_CAF1 80942 98 + 1 ----CGAAGGAGUAGAAGGACCUAGCAGGCCGCAUUUUAUGAAUUAUUCAAAAUGAUCAUAAUAAAUUGCUGCAGCCUUUUACGGCAGCUGGGAAAU---GGGAA---- ----................((((((..((.(((..((((((.((((.....)))))))))).....))).)).(((......))).))))))....---.....---- ( -25.10) >DroAna_CAF1 91833 107 + 1 GUUACGAAGGAAGCCGACGACCUAGCAGGCCGCAUUUUAUGAAUUAUUCAAAAUGAUCAUAAUAAAUUGCUGCAGCCUUUUACGGCAGCUGGGAACAA--GGGGACUUG ....((..((...))..)).((((((..((.(((..((((((.((((.....)))))))))).....))).)).(((......))).))))))..(((--(....)))) ( -32.30) >consensus ____CGAAGGAGUGGAAGGACCUAGCAGGCCGCAUUUUAUGAAUUAUUCAAAAUGAUCAUAAUAAAUUGCUGCAGCCUUUUACGGCAGCUGGGAAAU___GGGGA____ ....................((((((..((.(((..((((((.((((.....)))))))))).....))).)).(((......))).))))))................ (-24.04 = -24.23 + 0.19)


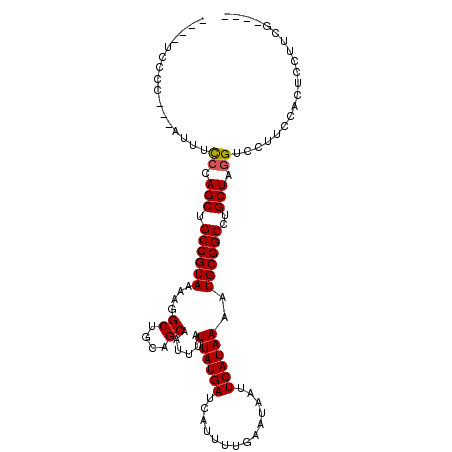
| Location | 26,774,374 – 26,774,475 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 91.37 |
| Mean single sequence MFE | -21.69 |
| Consensus MFE | -18.80 |
| Energy contribution | -18.82 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26774374 101 - 27905053 ----UCCCCCCUUUUUCCCAGCUGCCGUAAAAGGCUGCAGCAAUUUAUUAUGAUCAUUUUGAAUAAUUCAUAAAAUGCGGCCUGCUGGGUCCUUCCACUCCUUCG---- ----............((((((.((((((....((....))......((((((..............))))))..))))))..))))))................---- ( -26.04) >DroSec_CAF1 77774 98 - 1 ----UCCCC---AUUUCCCAGCUGCCGUAAAAGGCUGCAGCAAUUUAUUAUGAUCAUUUUGAAUAAUUCAUAAAAUGCGGCCUGCUAGGUCCUUCCACUCCUUCG---- ----.....---....((.(((.((((((....((....))......((((((..............))))))..))))))..))).))................---- ( -19.54) >DroSim_CAF1 81483 98 - 1 ----UCCCC---AUUUCCCAGCUGCCGUAAAAGGCUGCAGCAAUUUAUUAUGAUCAUUUUGAAUAAUUCAUAAAAUGCGGCCUGCUAGGUCCUUCCACUCCUUCG---- ----.....---....((.(((.((((((....((....))......((((((..............))))))..))))))..))).))................---- ( -19.54) >DroEre_CAF1 79301 98 - 1 ----GCCCC---AUUUUGCAGCUGCCGUAAAAGGCUGCAGCAAUUUAUUAUGAUCAUUUUGAAUAAUUCAUAAAAUGCGGCCUGCUAGGUCCUUCUACUCCUUCG---- ----(((.(---(((((((.((.(((......))).)).))))....((((((..............)))))))))).)))....((((....))))........---- ( -21.64) >DroYak_CAF1 80942 98 - 1 ----UUCCC---AUUUCCCAGCUGCCGUAAAAGGCUGCAGCAAUUUAUUAUGAUCAUUUUGAAUAAUUCAUAAAAUGCGGCCUGCUAGGUCCUUCUACUCCUUCG---- ----.....---....((.(((.((((((....((....))......((((((..............))))))..))))))..))).))................---- ( -19.54) >DroAna_CAF1 91833 107 - 1 CAAGUCCCC--UUGUUCCCAGCUGCCGUAAAAGGCUGCAGCAAUUUAUUAUGAUCAUUUUGAAUAAUUCAUAAAAUGCGGCCUGCUAGGUCGUCGGCUUCCUUCGUAAC ((((....)--)))...((.((.(((.....((((((((........((((((..............))))))..))))))))....))).)).))............. ( -23.84) >consensus ____UCCCC___AUUUCCCAGCUGCCGUAAAAGGCUGCAGCAAUUUAUUAUGAUCAUUUUGAAUAAUUCAUAAAAUGCGGCCUGCUAGGUCCUUCCACUCCUUCG____ ................((.(((.((((((....((....))......((((((..............))))))..))))))..))).)).................... (-18.80 = -18.82 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:46 2006