| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 613,531 – 613,641 |
| Length | 110 |
| Max. P | 0.582990 |

| Location | 613,531 – 613,641 |
|---|---|
| Length | 110 |
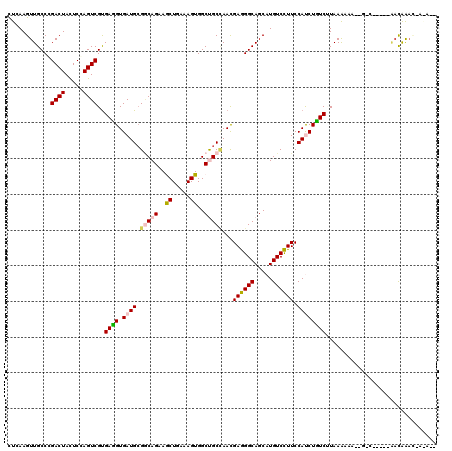
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.33 |
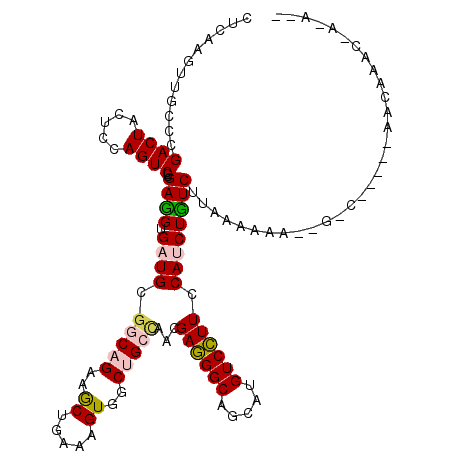
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -21.24 |
| Energy contribution | -21.18 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 613531 110 - 27905053 CUAAAGUUGCCCGACUACUCUAGUCGCGAGGUGAUGAGGCAGAAGCUGAAAGUGGCUGCUAACGAAGGCAGCAUGUCUUUCCACCUCUCAUAAGCAA--G-C-----AACAAACCAAA-- .....(((((.((((((...)))))).((((((..((((((...((.....)).((((((......)))))).))))))..))))))..........--)-)-----)))........-- ( -38.90) >DroVir_CAF1 46515 106 - 1 CUCAAGUUGCCGGACUACUCCAGUCGUGAUGUGAUGCGACAGAAGCUGAAAGUGGCUGCCAACGAGGGCAGCAUGUCCUUCCAUCUGUCUUAAAAAG--AAC-----AACAAA------- .....((((..((((.......(((((........)))))..............((((((......))))))..)))).....(((.........))--).)-----)))...------- ( -28.00) >DroGri_CAF1 39741 115 - 1 CUCAAGUUGCCGGACUACUCCAGUCGUGAUGUGAUGCGGCAGAAACUGAAAGUGGCUGCCAAUGAGGGCAGCAUGUCCUUCCAUCUGUCUUAAAACU--UAAUCUUACACAA---AGCAA .(((.((..(..((((.....)))))..)).)))((((((((..((.....))..)))))....(((((((.(((......))))))))))......--.............---.))). ( -30.70) >DroYak_CAF1 38661 110 - 1 CUAAAGUUGCCAGACUACUCCAGUCGUGAGGUGAUGCGCCAGAAGCUGAAAGUGGCCGCAAACGAGGGCAGCAUGUCCUUCCACCUCUCAUAAGGAA--G-U-----AUCGGACCAAA-- .........((.((.((((((....(.((((((.(((((((...........))).))))...((((((.....)))))).)))))).)....)).)--)-)-----)))))......-- ( -31.20) >DroMoj_CAF1 40052 113 - 1 CUCAAGUUGCCCGACUACUCGAGUCGUGAUGUGAUGCGGCAGAAGCUGAAAGUGGCUGCCAACGAGGGCAGCAUGUCCUUCCAUCUGUCUUAAAAAG--AAC-----AACAAAUUAGAAA .....((((..(((((.....))))).((((.((((.(((((..((.....))..)))))...((((((.....)))))).))))))))........--..)-----))).......... ( -31.70) >DroAna_CAF1 34924 108 - 1 UUGAAGUUGCCCGACUAUUCGAGUCGUGAGGUGAUGCGGCAGAAACUAAAAGUGGCCGCUAACGAGGGCAGCAUGUCCUUCCAUCUUUCCUAAACCAAUG-C-----GAUAAAC------ .....(((((.(((((.....))))).((((((..(((((....((.....)).)))))....((((((.....)))))).))))))............)-)-----)))....------ ( -31.20) >consensus CUCAAGUUGCCCGACUACUCCAGUCGUGAGGUGAUGCGGCAGAAGCUGAAAGUGGCUGCCAACGAGGGCAGCAUGUCCUUCCAUCUGUCUUAAAAAA__G_C_____AACAAAC_A_A__ ............((((.....))))..((((.((((.(((((..((.....))..)))))...((((((.....)))))).))))))))............................... (-21.24 = -21.18 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:05 2006