| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 138,643 – 138,748 |
| Length | 105 |
| Max. P | 0.646443 |

| Location | 138,643 – 138,748 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -20.90 |
| Energy contribution | -22.12 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
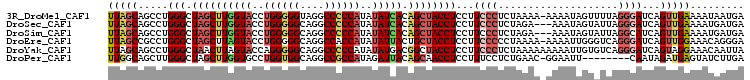
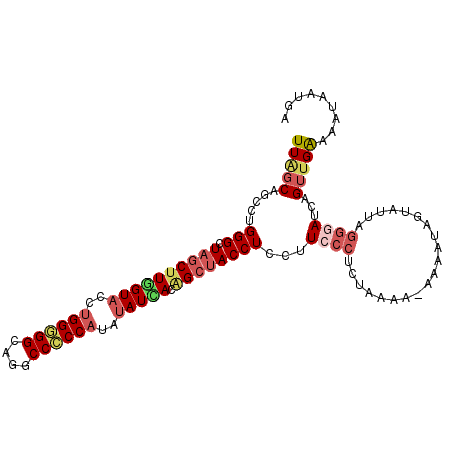
>3R_DroMel_CAF1 138643 105 + 27905053 UCAUUAUUUUCAACUGAUCCCUAAAACUAUUUU-UUUUAGAGGGAAGGAGGUAGCUGUGAUAUAUGGGGGCCUACCCCCAGGUACCAAGCUAGCCCAGGCUGCUAA .................(((((....(((....-...)))))))).((.(.(((((.((.(((.((((((....)))))).))).))))))).))).......... ( -30.70) >DroSec_CAF1 1267 103 + 1 UCAUCAUUUUCAACUGAUCCCUAAUACUAUUU---UCUAGAGGGAAGGAGGUAGCUGUGAUAUAUGGGGGCCUGCCCCCAGGUACCAAGCUAGCCCAGGCUGCUAA .................(((((....(((...---..)))))))).((.(.(((((.((.(((.((((((....)))))).))).))))))).))).......... ( -31.50) >DroSim_CAF1 2221 103 + 1 UCAUCAUUUUCAACUGAUGCCUAAUACUAUUU---UCUAGAGGGAAGGAGGUAGCUGUGAUAUAUGGGGGCCUGCCCCCAGGUACCAAGCUAGCCCAGGCUGCUAA ..((((........))))((((.......((.---.(....)..))((.(.(((((.((.(((.((((((....)))))).))).))))))).)))))))...... ( -33.60) >DroEre_CAF1 2152 105 + 1 UCCCUGUUUCCAACUGAUCCCUGACCCAAUUUU-UUUUAGGGGGAAGGAGGUAGCAGUAAUAUAUGGUGGCCUGCCCCCAGGUACUAAGCUAGCCCAGGCGGCUAA ...(((((.((..((..(((((((.........-..)))))))..))..)).))))).......(((((.((((....)))))))))((((.((....)))))).. ( -33.00) >DroYak_CAF1 2411 106 + 1 UAAUUGUUUCCUACUGAUCCCUGACACAAUUUUUUUUUAGAGGGAAGGAGGUAGCCGUCAUAUAUGGGGGCCUGCCCCCUGGUACUAAGUUAGCCCAGGCUGCUAA ...((.((((((.((((...................)))))))))).))(((((((.........(((((....))))).(((.........)))..))))))).. ( -32.41) >DroPer_CAF1 1444 97 + 1 UCAAGAUACUCAUCUAUUG--------AAUUCC-GUUCAGAGGAAAGGAGGUUGCUGUAAUCUAUGGCGGCCUGCCACCAGGCACCAAGCUAGCCCAAGCUGCCAA (((((((....))))..))--------).((((-.......)))).(((((((((((((...))))))))))).))....((((....(((......))))))).. ( -27.30) >consensus UCAUUAUUUUCAACUGAUCCCUAACACAAUUUU_UUUUAGAGGGAAGGAGGUAGCUGUGAUAUAUGGGGGCCUGCCCCCAGGUACCAAGCUAGCCCAGGCUGCUAA .............((..(((((..................))))).)).(((((((........((((((....))))))(((.........)))..))))))).. (-20.90 = -22.12 + 1.22)

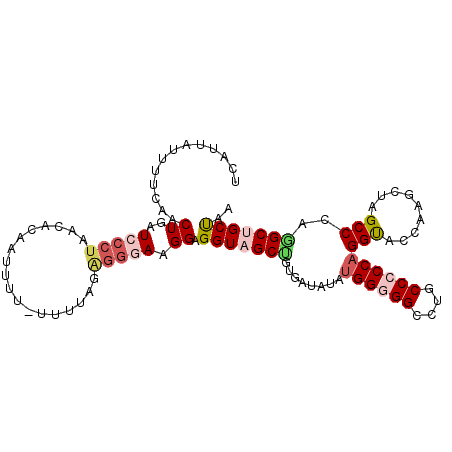
| Location | 138,643 – 138,748 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 81.43 |
| Mean single sequence MFE | -31.27 |
| Consensus MFE | -21.27 |
| Energy contribution | -22.50 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646443 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 138643 105 - 27905053 UUAGCAGCCUGGGCUAGCUUGGUACCUGGGGGUAGGCCCCCAUAUAUCACAGCUACCUCCUUCCCUCUAAAA-AAAAUAGUUUUAGGGAUCAGUUGAAAAUAAUGA ....((((.((((.((((((((((..((((((....))))))..))))).))))))).......((((((((-.......))))))))..)))))).......... ( -34.10) >DroSec_CAF1 1267 103 - 1 UUAGCAGCCUGGGCUAGCUUGGUACCUGGGGGCAGGCCCCCAUAUAUCACAGCUACCUCCUUCCCUCUAGA---AAAUAGUAUUAGGGAUCAGUUGAAAAUGAUGA ....((((.((((.((((((((((..((((((....))))))..))))).)))))))....((((((((..---...)))....))))).)))))).......... ( -35.00) >DroSim_CAF1 2221 103 - 1 UUAGCAGCCUGGGCUAGCUUGGUACCUGGGGGCAGGCCCCCAUAUAUCACAGCUACCUCCUUCCCUCUAGA---AAAUAGUAUUAGGCAUCAGUUGAAAAUGAUGA ......(((((((.((((((((((..((((((....))))))..))))).))))))).........(((..---...)))...)))))((((........)))).. ( -33.90) >DroEre_CAF1 2152 105 - 1 UUAGCCGCCUGGGCUAGCUUAGUACCUGGGGGCAGGCCACCAUAUAUUACUGCUACCUCCUUCCCCCUAAAA-AAAAUUGGGUCAGGGAUCAGUUGGAAACAGGGA .......((((..((((((........((..((((..............))))..))(((((..(((.....-......)))..)))))..))))))...)))).. ( -31.44) >DroYak_CAF1 2411 106 - 1 UUAGCAGCCUGGGCUAACUUAGUACCAGGGGGCAGGCCCCCAUAUAUGACGGCUACCUCCUUCCCUCUAAAAAAAAAUUGUGUCAGGGAUCAGUAGGAAACAAUUA .....(((((((((((...)))).)))(((((....))))).........)))).(((.(((((((..................)))))..)).)))......... ( -29.57) >DroPer_CAF1 1444 97 - 1 UUGGCAGCUUGGGCUAGCUUGGUGCCUGGUGGCAGGCCGCCAUAGAUUACAGCAACCUCCUUUCCUCUGAAC-GGAAUU--------CAAUAGAUGAGUAUCUUGA ..((((((....))).)))(((((((((....)))).))))).((((..........(((.(((....))).-)))(((--------((.....)))))))))... ( -23.60) >consensus UUAGCAGCCUGGGCUAGCUUGGUACCUGGGGGCAGGCCCCCAUAUAUCACAGCUACCUCCUUCCCUCUAAAA_AAAAUAGUAUUAGGGAUCAGUUGAAAAUAAUGA (((((.....(((.((((((((((..((((((....))))))..))))).))))))))...((((....................))))...)))))......... (-21.27 = -22.50 + 1.23)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:09 2006