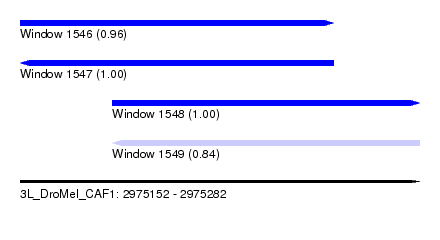
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,975,152 – 2,975,282 |
| Length | 130 |
| Max. P | 0.999483 |

| Location | 2,975,152 – 2,975,254 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -21.76 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.74 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2975152 102 + 23771897 AUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUUGUGCUGUAUGGACGGGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCA ..(((.((((....)))).....(((((.......)))))(((((((...(((((.(((((.............))))).)))))..)))))))...))).. ( -23.92) >DroSec_CAF1 9418 100 + 1 AUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUU--UCUGUAUGGACGGGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCA ..(((((((....(((((......)))))(((....--..)))))))((((((...(((((.............)))))......))))))......))).. ( -21.62) >DroSim_CAF1 9361 100 + 1 AUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUU--UGUGUAUGGACGAGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCA ..(((........(((((......)))))......(--(((((((((...(((((.(((((.............))))).)))))..))))))))))))).. ( -24.72) >DroEre_CAF1 11690 100 + 1 AUGGGCCAUAAAAAUAUAAAAACGCAUAUACAAAUU--UCUGUAUGGAUGGGUUUACAGCUGAAGUGUUUCAAAAGCUGAAAACUACCCGUAUAUAAGCUCA .(((((.......((((........)))).......--..(((((((...(((((.(((((.............))))).)))))..)))))))...))))) ( -21.42) >DroYak_CAF1 9570 100 + 1 AUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUU--UCUGUAUGAAUGGGUUUACAGCUGAAAUGUUUCAAAGGCUGCAAACUAUCCGUAUAUAAGCUCG ..(((........(((((......))))).......--..((((((....(((((.((((((((....)))...))))).)))))...))))))...))).. ( -17.10) >consensus AUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUU__UCUGUAUGGACGGGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCA ..(((........(((((......)))))...........(((((((...(((((.(((((.............))))).)))))..)))))))...))).. (-18.46 = -18.74 + 0.28)

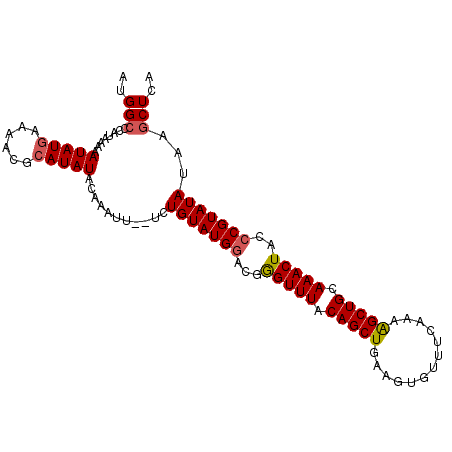
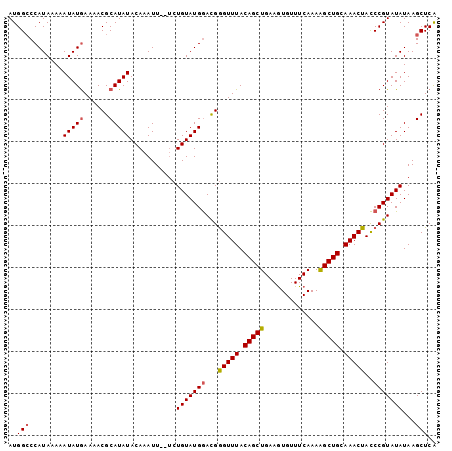
| Location | 2,975,152 – 2,975,254 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.25 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -22.00 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2975152 102 - 23771897 UGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACCCGUCCAUACAGCACAAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAU ((.(((((((.((((((...((((((((...(((....)))))))))))))))))..((((((......)))))).................))))))))). ( -27.00) >DroSec_CAF1 9418 100 - 1 UGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACCCGUCCAUACAGA--AAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAU ((.(((((((.((((((...((((((((...(((....)))))))))))))))))..((((((.--...)))))).................))))))))). ( -26.90) >DroSim_CAF1 9361 100 - 1 UGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACUCGUCCAUACACA--AAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAU .((((.((((((((((((((((((((((...(((....)))))))))))))).((......)).--.))))))))))).))))(((((....)))))..... ( -25.30) >DroEre_CAF1 11690 100 - 1 UGAGCUUAUAUACGGGUAGUUUUCAGCUUUUGAAACACUUCAGCUGUAAACCCAUCCAUACAGA--AAUUUGUAUAUGCGUUUUUAUAUUUUUAUGGCCCAU .((((.(((((((((((.((((.(((((...(((....)))))))).))))..))))....(..--..)..))))))).))))................... ( -18.40) >DroYak_CAF1 9570 100 - 1 CGAGCUUAUAUACGGAUAGUUUGCAGCCUUUGAAACAUUUCAGCUGUAAACCCAUUCAUACAGA--AAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAU .((((.(((((((((((.(((((((((...((((....)))))))))))))...(((.....))--)))))))))))).))))(((((....)))))..... ( -25.70) >consensus UGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACCCGUCCAUACAGA__AAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAU .(.(((((((...((((.(((((((((...((((....)))))))))))))..))))(((((((....))))))).................)))))))).. (-22.00 = -22.24 + 0.24)

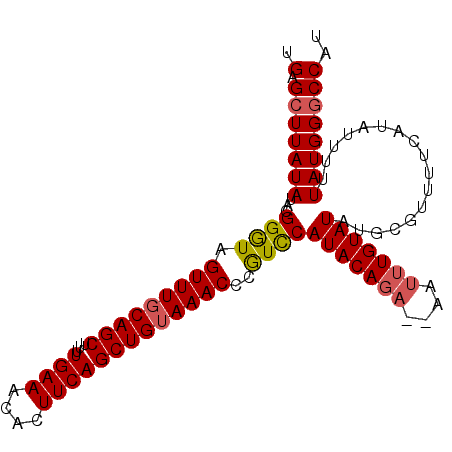
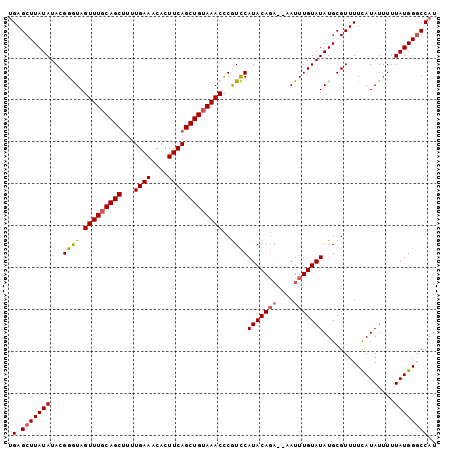
| Location | 2,975,182 – 2,975,282 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -26.92 |
| Energy contribution | -27.04 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2975182 100 + 23771897 CAAAUUGUGCUGUAUGGACGGGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCAAUGGAUUCUGCCGAUUGGCUUAAAGGCU ........(((((((((...(((((.(((((.............))))).)))))..)))))).(((((.((((((......)).)))))))))..))). ( -28.62) >DroSec_CAF1 9448 98 + 1 CAAAUU--UCUGUAUGGACGGGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCAUUGGAUUCUGCCGAUUGGCUUAAAGGCU ......--..(((((((...(((((.(((((.............))))).)))))..)))))))((((((.(((((......))))).))))))...... ( -29.12) >DroSim_CAF1 9391 98 + 1 CAAAUU--UGUGUAUGGACGAGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCAUUGGAUUCUGCCGAUUGGCUUAAAGGCU .....(--(((((((((...(((((.(((((.............))))).)))))..))))))))))(((.(((((......))))).)))......... ( -29.22) >DroEre_CAF1 11720 98 + 1 CAAAUU--UCUGUAUGGAUGGGUUUACAGCUGAAGUGUUUCAAAAGCUGAAAACUACCCGUAUAUAAGCUCAUUCGAUUCUGCCGAUUGGCUUAAAGGCU ......--..(((((((...(((((.(((((.............))))).)))))..)))))))(((((.((.(((.......))).)))))))...... ( -26.52) >DroYak_CAF1 9600 98 + 1 CAAAUU--UCUGUAUGAAUGGGUUUACAGCUGAAAUGUUUCAAAGGCUGCAAACUAUCCGUAUAUAAGCUCGUUGGAUGCUGCCGAUUGGCUUAAAGGCU ......--..((((((....(((((.((((((((....)))...))))).)))))...))))))((((((.(((((......))))).))))))...... ( -25.40) >consensus CAAAUU__UCUGUAUGGACGGGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCAUUGGAUUCUGCCGAUUGGCUUAAAGGCU ..........(((((((...(((((.(((((.............))))).)))))..)))))))((((((.(((((......))))).))))))...... (-26.92 = -27.04 + 0.12)

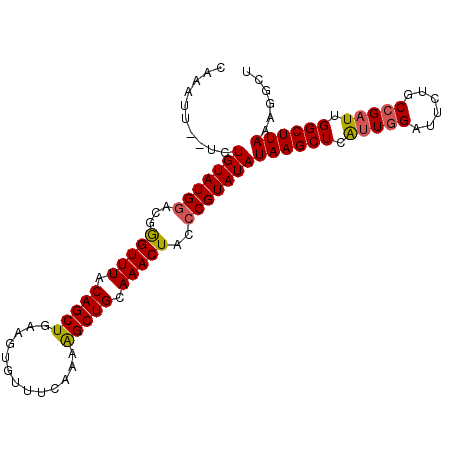
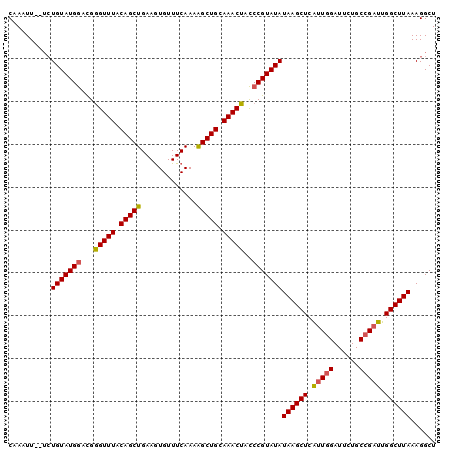
| Location | 2,975,182 – 2,975,282 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 93.72 |
| Mean single sequence MFE | -23.34 |
| Consensus MFE | -18.98 |
| Energy contribution | -18.62 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2975182 100 - 23771897 AGCCUUUAAGCCAAUCGGCAGAAUCCAUUGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACCCGUCCAUACAGCACAAUUUG .........(((....))).......((((.((((((..((((((...((((((((...(((....)))))))))))))))))..))).))).))))... ( -28.40) >DroSec_CAF1 9448 98 - 1 AGCCUUUAAGCCAAUCGGCAGAAUCCAAUGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACCCGUCCAUACAGA--AAUUUG .........(((....)))......(((....(((((..((((((...((((((((...(((....)))))))))))))))))..))).)).--...))) ( -23.50) >DroSim_CAF1 9391 98 - 1 AGCCUUUAAGCCAAUCGGCAGAAUCCAAUGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACUCGUCCAUACACA--AAUUUG .((((.(((((..((.((......)).))..))))).....))))(((((((((((...(((....))))))))))))))............--...... ( -22.50) >DroEre_CAF1 11720 98 - 1 AGCCUUUAAGCCAAUCGGCAGAAUCGAAUGAGCUUAUAUACGGGUAGUUUUCAGCUUUUGAAACACUUCAGCUGUAAACCCAUCCAUACAGA--AAUUUG ......(((((((.((((.....)))).)).))))).....((((.((((.(((((...(((....)))))))).))))..)))).......--...... ( -21.50) >DroYak_CAF1 9600 98 - 1 AGCCUUUAAGCCAAUCGGCAGCAUCCAACGAGCUUAUAUACGGAUAGUUUGCAGCCUUUGAAACAUUUCAGCUGUAAACCCAUUCAUACAGA--AAUUUG .........(((....)))(((.((....))))).......((((.(((((((((...((((....)))))))))))))..)))).......--...... ( -20.80) >consensus AGCCUUUAAGCCAAUCGGCAGAAUCCAAUGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACCCGUCCAUACAGA__AAUUUG .........(((....)))......................((((.(((((((((...((((....)))))))))))))..))))............... (-18.98 = -18.62 + -0.36)

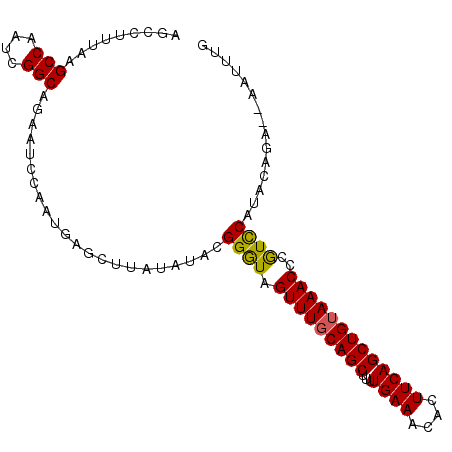
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:34 2006