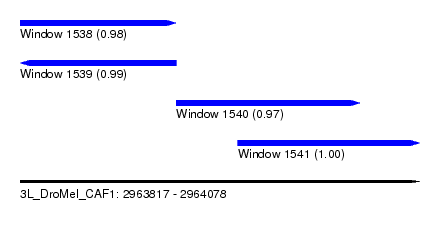
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,963,817 – 2,964,078 |
| Length | 261 |
| Max. P | 0.997693 |

| Location | 2,963,817 – 2,963,919 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -35.22 |
| Consensus MFE | -27.50 |
| Energy contribution | -28.26 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2963817 102 + 23771897 CAAUUCCUCAAAUUAAGUAAAGUGAGUAGGGGAAUCCCCUUUUCCAUUUAAAUUUGCAUAAGAGGCAUCGGCGAAAGC------GGAUGAGGAUGCGGAUGGGUUGCA ((((((...........((((..(((.(((((...))))).)))..)))).((((((((......((((.((....))------.))))...)))))))))))))).. ( -31.40) >DroSec_CAF1 192460 108 + 1 CAAUUCCUCAAAUUAAGUAAAGUGAGUAGGGGAAUCCCCUUUUCCAUUUAAAUUUGCAUAAGAGGCAUCGGCGAAUGCGGAUGCGGAUGAGGAUGCGGAUGGGUUGCA ....((((((......(((((..(((.(((((...))))).)))........))))).......(((((.((....)).)))))...))))))(((((.....))))) ( -31.20) >DroSim_CAF1 178569 108 + 1 CAAUUCCUCAAAUUAAGUAAAGUGAGUAGGGGAAUCCCCUUUUCCAUUUAAAUUUGCAUAAGAGGCAUCGGCGAAUGCGGAUGCGGAUGAGGAUGCGGAUGGGUUGCA ....((((((......(((((..(((.(((((...))))).)))........))))).......(((((.((....)).)))))...))))))(((((.....))))) ( -31.20) >DroEre_CAF1 200551 108 + 1 CAAUUCCUCAAAUUAAGCAAAGUGGAUAGGGGAAUCCCCUUUUCCAUUUAAAUUUGCAUAAGAGGCAUCGGCGAAGGCGGAUGCCGUUGAGGAUGCGGAUGGGUUGCA ....(((((((.....((((((((((.(((((....))))).))))).....)))))......((((((.((....)).)))))).)))))))(((((.....))))) ( -45.90) >DroYak_CAF1 204769 108 + 1 CAAUACCUCAAAUUAAGUAAAGUGGAUAGGGGAAUCCCCUUUUCCAUUUAAAUUUGCAUAAGAGGCAUCGACGGAUGCGGAUGCGGAUGAGGAUGCGGAUGGGUUGCA .....((((..........(((((((.(((((....))))).)))))))..((((((((.....(((((....)))))..)))))))))))).(((((.....))))) ( -36.40) >consensus CAAUUCCUCAAAUUAAGUAAAGUGAGUAGGGGAAUCCCCUUUUCCAUUUAAAUUUGCAUAAGAGGCAUCGGCGAAUGCGGAUGCGGAUGAGGAUGCGGAUGGGUUGCA ....((((((......(((((..(((.(((((...))))).)))........))))).......(((((.((....)).)))))...))))))(((((.....))))) (-27.50 = -28.26 + 0.76)

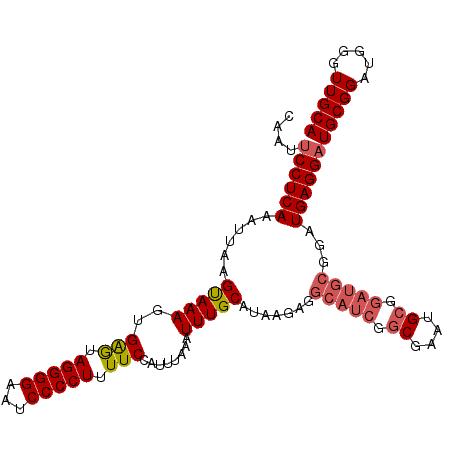
| Location | 2,963,817 – 2,963,919 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 93.80 |
| Mean single sequence MFE | -27.04 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2963817 102 - 23771897 UGCAACCCAUCCGCAUCCUCAUCC------GCUUUCGCCGAUGCCUCUUAUGCAAAUUUAAAUGGAAAAGGGGAUUCCCCUACUCACUUUACUUAAUUUGAGGAAUUG (((.........)))(((((((((------((....))...(((.......))).........)))..(((((...))))).................)))))).... ( -20.80) >DroSec_CAF1 192460 108 - 1 UGCAACCCAUCCGCAUCCUCAUCCGCAUCCGCAUUCGCCGAUGCCUCUUAUGCAAAUUUAAAUGGAAAAGGGGAUUCCCCUACUCACUUUACUUAAUUUGAGGAAUUG (((.........)))((((((((((((((.((....)).)))))...(((........)))..)))..(((((...))))).................)))))).... ( -26.60) >DroSim_CAF1 178569 108 - 1 UGCAACCCAUCCGCAUCCUCAUCCGCAUCCGCAUUCGCCGAUGCCUCUUAUGCAAAUUUAAAUGGAAAAGGGGAUUCCCCUACUCACUUUACUUAAUUUGAGGAAUUG (((.........)))((((((((((((((.((....)).)))))...(((........)))..)))..(((((...))))).................)))))).... ( -26.60) >DroEre_CAF1 200551 108 - 1 UGCAACCCAUCCGCAUCCUCAACGGCAUCCGCCUUCGCCGAUGCCUCUUAUGCAAAUUUAAAUGGAAAAGGGGAUUCCCCUAUCCACUUUGCUUAAUUUGAGGAAUUG (((.........)))(((((((.((((((.((....)).))))))..(((.(((((......((((..(((((...))))).)))).))))).))).))))))).... ( -34.90) >DroYak_CAF1 204769 108 - 1 UGCAACCCAUCCGCAUCCUCAUCCGCAUCCGCAUCCGUCGAUGCCUCUUAUGCAAAUUUAAAUGGAAAAGGGGAUUCCCCUAUCCACUUUACUUAAUUUGAGGUAUUG (((.........))).(((((...((((..(((((....))))).....)))).....((((((((..(((((...))))).)))).)))).......)))))..... ( -26.30) >consensus UGCAACCCAUCCGCAUCCUCAUCCGCAUCCGCAUUCGCCGAUGCCUCUUAUGCAAAUUUAAAUGGAAAAGGGGAUUCCCCUACUCACUUUACUUAAUUUGAGGAAUUG (((.........)))((((((...(((((.((....)).)))))..............(((((((...(((((...)))))..))).)))).......)))))).... (-21.96 = -22.60 + 0.64)

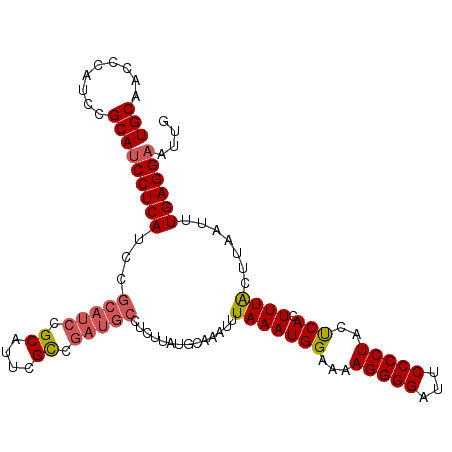
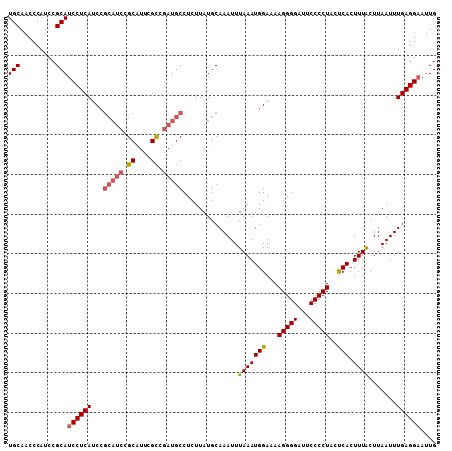
| Location | 2,963,919 – 2,964,039 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -28.86 |
| Energy contribution | -28.86 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2963919 120 + 23771897 GUGUAAUUGGGCGAAGUGAAAUUGCGUUAAUAUUUAACAAGCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGCGGAAAAGGGAUACGGAAACGUAGUCCUUCGGGGCAAAGUGCGAA .(((..((((((...((......))((((.....))))..))).((....)).)))..)))...((((((................((((....))))((((....))))..)))))).. ( -32.40) >DroSec_CAF1 192568 120 + 1 GUGUAAUUGGGCGAAGUGAAAUUGCGUUAAUAUUUAACAAGCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGCGGAAAAGGGAUACGGAAACGUCGUCCUUCGGGGCAAAGUGCGAA .(((..((((((...((......))((((.....))))..))).((....)).)))..)))...((((((...(((.....(((((((((....))).))))))....))).)))))).. ( -31.00) >DroSim_CAF1 178677 120 + 1 GUGUAAUUGGGCGAAGUGAAAUUGCGUUAAUAUUUAACAAGCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGCGGAAAAGGGAUACGGAAACGUCGUCCUUCGGGGCAAAGUGCGAA .(((..((((((...((......))((((.....))))..))).((....)).)))..)))...((((((...(((.....(((((((((....))).))))))....))).)))))).. ( -31.00) >DroEre_CAF1 200659 120 + 1 GUGUAAUUGGGCGAAGUGAAAUUGCGUUAAUAUUUAACAAGCCGGCGAAAGUGCAAAAGCGAAGCGCAUCAAAUGGGGAAAAGGGAUACGGAAGCGUCGUCCUUCGGGGCAAAGUGCGAA ......(((.((((.(((...((((((((.....))))..((..((....))))....))))..))).))..............((..(....)..))((((....))))...)).))). ( -30.30) >DroYak_CAF1 204877 120 + 1 GUGUAAUUGGGCGAAGUGAAAUUGCGUUAAUAUUUAACAAGCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGGGCAAAAGGGAUACGGAAACGUCGUCCUUCGGGGCAAAGUGCGAA ......(((.((...(((...((((((((.....))))..((..((....))))....))))..))).........((...(((((((((....))).))))))....))...)).))). ( -30.80) >consensus GUGUAAUUGGGCGAAGUGAAAUUGCGUUAAUAUUUAACAAGCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGCGGAAAAGGGAUACGGAAACGUCGUCCUUCGGGGCAAAGUGCGAA ......(((.((.............((((.....))))..(((.(((....(((....)))...)))..............(((((((((....))).))))))...)))...)).))). (-28.86 = -28.86 + -0.00)

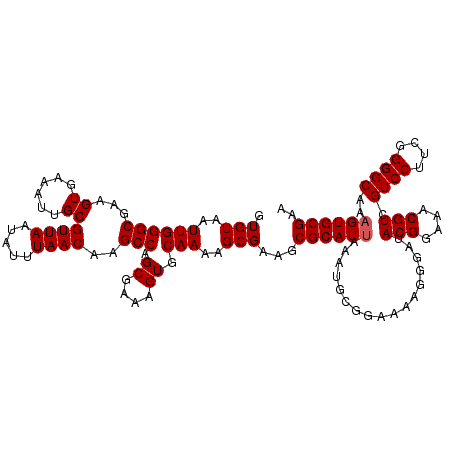
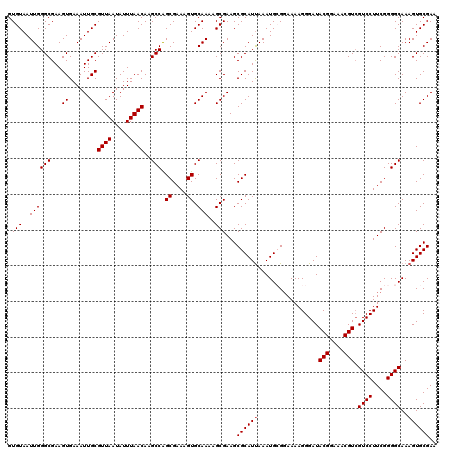
| Location | 2,963,959 – 2,964,078 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.40 |
| Mean single sequence MFE | -39.94 |
| Consensus MFE | -34.44 |
| Energy contribution | -34.60 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997693 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2963959 119 + 23771897 GCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGCGGAAAAGGGAUACGGAAACGUAGUCCUUCGGGGCAAAGUGCGAAAUUAUGCGGAGUUCAGGGAGAUCCGCA-AGCCCUUUCCUU ((..((....))((....))...))((((...))))(((((.(((.((((....))))((((....))))..............((((((.(((...))).))))))-..)))))))).. ( -42.20) >DroSec_CAF1 192608 119 + 1 GCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGCGGAAAAGGGAUACGGAAACGUCGUCCUUCGGGGCAAAGUGCGAAAUUAUGCGGAAUUCAGGGAGAUCCGCA-AACCCCUUGCUU ((..((....))))..((((....(((((...)))))...(((((..(((....))).((((....))))..............((((((.(((...))).))))))-...))))))))) ( -40.50) >DroSim_CAF1 178717 119 + 1 GCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGCGGAAAAGGGAUACGGAAACGUCGUCCUUCGGGGCAAAGUGCGAAAUUAUGCGGAAUUCAGGGAGAUCCGCA-AACCCUUUGCUU ((..((....))))..((((((((((((((...(((.....(((((((((....))).))))))....))).))))))......((((((.(((...))).))))))-....)))))))) ( -42.10) >DroEre_CAF1 200699 120 + 1 GCCGGCGAAAGUGCAAAAGCGAAGCGCAUCAAAUGGGGAAAAGGGAUACGGAAGCGUCGUCCUUCGGGGCAAAGUGCGAAAUUAUGCGGAAUUCAGGGAAAUCCGCAGAACCCUUUCCUU ....(((....(((....)))...))).......(((((((.(((..(((....))).((((....))))..............((((((.(((...))).))))))...)))))))))) ( -38.60) >DroYak_CAF1 204917 120 + 1 GCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGGGCAAAAGGGAUACGGAAACGUCGUCCUUCGGGGCAAAGUGCGAAUUUAUGCGGAAUUCAGGGCGAUCUGCAAAACCCUUUCCUU (((.((....))......(((...)))........))).((((((..(((....))).((((....))))..............((((((..((.....))))))))...)))))).... ( -36.30) >consensus GCCAGCGAAAGUGCAAAAGCGAAGCGCAUUAAAUGCGGAAAAGGGAUACGGAAACGUCGUCCUUCGGGGCAAAGUGCGAAAUUAUGCGGAAUUCAGGGAGAUCCGCA_AACCCUUUCCUU ....(((....(((....)))...))).......(((((..(((((((((....))).)))))).(((((.....)).......((((((.(((...))).))))))...)))))))).. (-34.44 = -34.60 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:26 2006