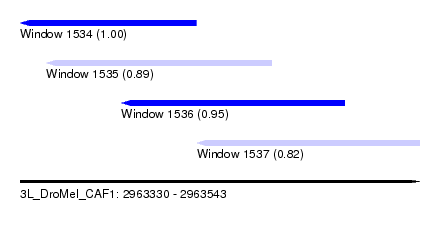
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,963,330 – 2,963,543 |
| Length | 213 |
| Max. P | 0.998392 |

| Location | 2,963,330 – 2,963,424 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 96.70 |
| Mean single sequence MFE | -26.14 |
| Consensus MFE | -24.96 |
| Energy contribution | -24.76 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.95 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2963330 94 - 23771897 AAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCGAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCCCCGCGCAUUUUAAAGAC ...((((((..(((.(((((((....)))))))))).((.(((((.....)))))(((((((.((...)).)))))))))..))))))...... ( -27.20) >DroSec_CAF1 191974 94 - 1 AAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCCCCUCGCAUUUUAAAGAG .((((.((((.(((.(((((((....))))))))))....(((((.....)))))(((((((.((...)).))))))).)))).))))...... ( -28.40) >DroSim_CAF1 178093 94 - 1 AAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCCCCUCGCAUUUUAAAGAC .((((.((((.(((.(((((((....))))))))))....(((((.....)))))(((((((.((...)).))))))).)))).))))...... ( -28.40) >DroEre_CAF1 200074 94 - 1 AAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCUGCUCGCAUUUUAAAGAG .....((..(((((.(((((((....))))))))))(((..((((.....)))))))........))..))..(((.....))).......... ( -23.90) >DroYak_CAF1 204290 94 - 1 AAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCUCCCCGUAUUUUAAAGAC .....((....(((.(((((((....))))))))))))..(((((.....)))))(((((((.((...)).)))))))................ ( -22.80) >consensus AAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCCCCUCGCAUUUUAAAGAC ...((((((..(((.(((((((....))))))))))....(((((.....)))))(((((((.((...)).)))))))....))))))...... (-24.96 = -24.76 + -0.20)

| Location | 2,963,344 – 2,963,464 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.75 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -32.70 |
| Energy contribution | -32.50 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.891001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2963344 120 - 23771897 CUGUUUAUCCACGGACCAAAAUCGCCAGGAUGCCAUUGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCGAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCCCCG ...........((((......(((((.....((....))......)))))..((.(((((((....))))))))))))).(((((.....)))))(((((((.((...)).))))))).. ( -34.90) >DroSec_CAF1 191988 120 - 1 CUGUUUAUCCACGGACCAAAAUCGCCAGGAUGCCAUGGUCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCCCCU ...((((((....(((((..(((.....)))....))))).....))))))(((.(((((((....))))))))))....(((((.....)))))(((((((.((...)).))))))).. ( -35.60) >DroSim_CAF1 178107 120 - 1 CUGUUUAUCCACGGACCGAAAUCGCCAGGAUGCCAUGGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCCCCU ............(((((......((((((...)).))))......)))...(((.(((((((....))))))))))))..(((((.....)))))(((((((.((...)).))))))).. ( -35.70) >DroEre_CAF1 200088 120 - 1 CUGUUUAUCCACGGACCGAAAUCGCCAGGAUGCCAUGGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCUGCU ....((((((..(((((......((((((...)).))))......)))...(((.(((((((....))))))))))))..(((((.....)))))))))))..((...((....)).)). ( -33.90) >DroYak_CAF1 204304 120 - 1 CUGUUUAUCUACGGACCAAAAUCGCCAGGAUGCCAUGGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCUCCC ((((......)))).............(((.((.(((((....((((..(.(((.(((((((....))))))))))(((..((((.....)))))))....)..))))))))).))))). ( -35.30) >consensus CUGUUUAUCCACGGACCAAAAUCGCCAGGAUGCCAUGGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAAAUCGAACAAAUCGAUGGGUAAUUGCUUUGCCAUUGCCCCU ............(((......(((((.....((....))......)))))..((.(((((((....))))))))))))..(((((.....)))))(((((((.((...)).))))))).. (-32.70 = -32.50 + -0.20)

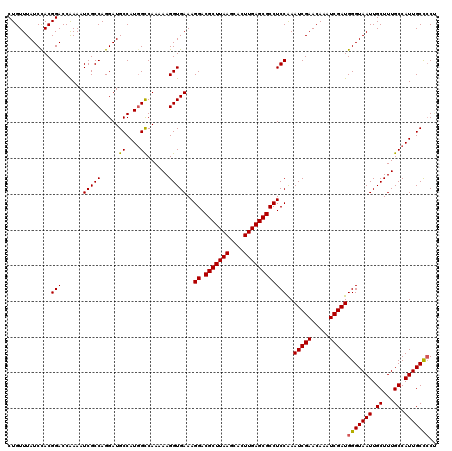
| Location | 2,963,384 – 2,963,503 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.99 |
| Mean single sequence MFE | -29.46 |
| Consensus MFE | -26.53 |
| Energy contribution | -26.41 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2963384 119 - 23771897 GGGCAAACAAAUAAACGCCACGCAAAUGCAAACAA-AACGCUGUUUAUCCACGGACCAAAAUCGCCAGGAUGCCAUUGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCGA ((..(((((.......((...))....((......-...)))))))..)).((((......(((((.....((....))......)))))..((.(((((((....))))))))))))). ( -28.80) >DroSec_CAF1 192028 119 - 1 GGGCAAACAAAUUAACGCCACGCAAAUGCAAACAA-AACGCUGUUUAUCCACGGACCAAAAUCGCCAGGAUGCCAUGGUCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAA ((.............((((..((....))(((((.-.....))))).......(((((..(((.....)))....))))).....))))..(((.(((((((....)))))))))))).. ( -30.30) >DroSim_CAF1 178147 118 - 1 G-GCAAACAAAUAAACGCCACGCAAAUGCAAACAA-AACGCUGUUUAUCCACGGACCGAAAUCGCCAGGAUGCCAUGGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAA (-((............)))..((....))(((((.-.....)))))......(((((......((((((...)).))))......)))...(((.(((((((....)))))))))))).. ( -29.20) >DroEre_CAF1 200128 120 - 1 GGGCAAACAAAUAAACGCCACGCAAAUGCAAACAAAAACGCUGUUUAUCCACGGACCGAAAUCGCCAGGAUGCCAUGGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAA ((.............((((..((....))..........(((((.(((((..((..((....)))).)))))..)))))......))))..(((.(((((((....)))))))))))).. ( -30.10) >DroYak_CAF1 204344 120 - 1 GGGCAAACAAAUAAACGCCACGCAAAUGCAAACAAAAACGCUGUUUAUCUACGGACCAAAAUCGCCAGGAUGCCAUGGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAA ((.............((((..((....))...........((((......)))).........((((((...)).))))......))))..(((.(((((((....)))))))))))).. ( -28.90) >consensus GGGCAAACAAAUAAACGCCACGCAAAUGCAAACAA_AACGCUGUUUAUCCACGGACCAAAAUCGCCAGGAUGCCAUGGCCAAAAAGGUGAAAGGACGCUUAAGCACUUGAGCGCCUCCAA ((..(((((............((....))............)))))..))..(((......(((((.....((....))......)))))..((.(((((((....)))))))))))).. (-26.53 = -26.41 + -0.12)

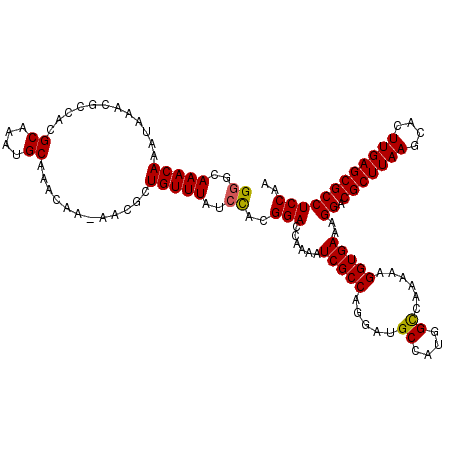
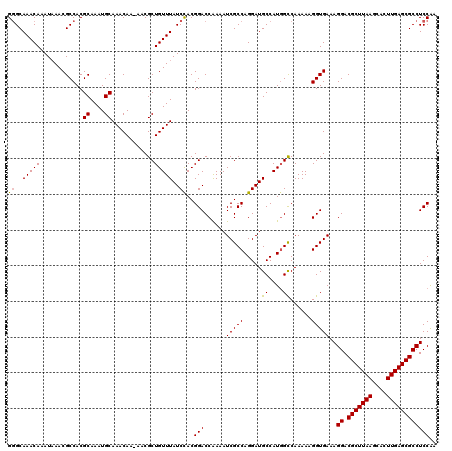
| Location | 2,963,424 – 2,963,543 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.34 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.816757 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2963424 119 - 23771897 GUCCAAGCUCCAAGCUCCUCGACCAGCAGACAUCAACAGCGGGCAAACAAAUAAACGCCACGCAAAUGCAAACAA-AACGCUGUUUAUCCACGGACCAAAAUCGCCAGGAUGCCAUUGCC (((..(((.....)))....)))..((((.....((((((((((............)))..((....))......-..)))))))(((((..((..........)).)))))...)))). ( -25.30) >DroSec_CAF1 192068 112 - 1 GUCCAA-------ACUCAUCGACCAGCAGACAUCAACAGCGGGCAAACAAAUUAACGCCACGCAAAUGCAAACAA-AACGCUGUUUAUCCACGGACCAAAAUCGCCAGGAUGCCAUGGUC ......-------.......((((((((..(...((((((((((............)))..((....))......-..))))))).......((..........)).)..)))..))))) ( -26.00) >DroSim_CAF1 178187 111 - 1 GUCCAA-------ACUCCUCGACCAGCAGACAUCAACAGCG-GCAAACAAAUAAACGCCACGCAAAUGCAAACAA-AACGCUGUUUAUCCACGGACCGAAAUCGCCAGGAUGCCAUGGCC ((((..-------.((.((.....)).)).....(((((((-((............)))..((....))......-...)))))).......)))).......((((((...)).)))). ( -24.20) >DroEre_CAF1 200168 113 - 1 UCCCAA-------ACUCCUCCACCAGCAGACAUCAGCAGCGGGCAAACAAAUAAACGCCACGCAAAUGCAAACAAAAACGCUGUUUAUCCACGGACCGAAAUCGCCAGGAUGCCAUGGCC ......-------.........((((((..(...((((((((((............)))..((....)).........))))))).......((..((....)))).)..)))..))).. ( -23.30) >DroYak_CAF1 204384 113 - 1 GCCCAA-------AUUCUGCGACCAGCAGACAUCAACAGCGGGCAAACAAAUAAACGCCACGCAAAUGCAAACAAAAACGCUGUUUAUCUACGGACCAAAAUCGCCAGGAUGCCAUGGCC (((...-------..(((((.....)))))((((((((((((((............)))..((....)).........)))))))..((....)).............))))....))). ( -30.50) >consensus GUCCAA_______ACUCCUCGACCAGCAGACAUCAACAGCGGGCAAACAAAUAAACGCCACGCAAAUGCAAACAA_AACGCUGUUUAUCCACGGACCAAAAUCGCCAGGAUGCCAUGGCC ((((........................(....)((((((((((............)))..((....)).........))))))).......)))).......((((((...)).)))). (-19.66 = -20.34 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:23 2006