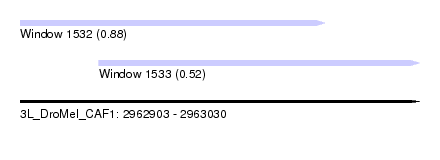
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,962,903 – 2,963,030 |
| Length | 127 |
| Max. P | 0.875101 |

| Location | 2,962,903 – 2,963,000 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -20.63 |
| Energy contribution | -21.78 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
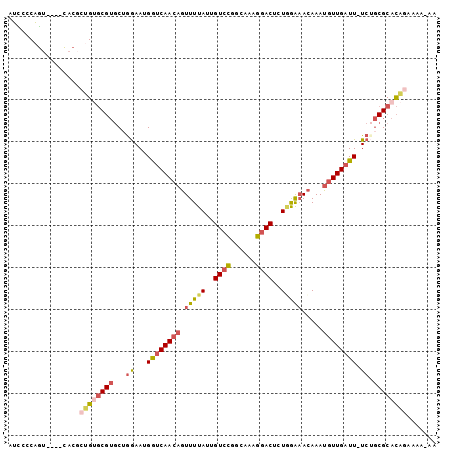
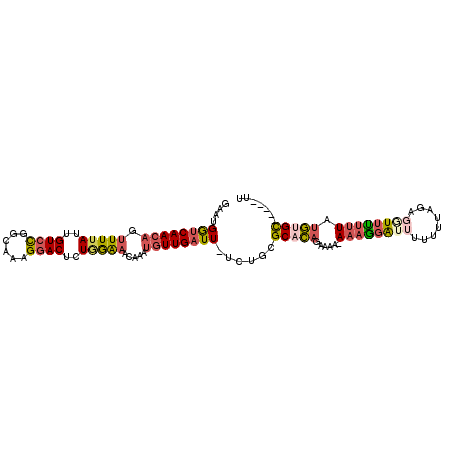
>3L_DroMel_CAF1 2962903 97 + 23771897 AUCCCCAGU----CACGCUGUGCGUGCUAGAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUGGAAACAAAUGUUGAUU-UCUGCGCACAGAAAAGAA .........----....((((((((...((((..((((((((((((...((((......))))....)))))...))))))))-)))))))))))....... ( -31.30) >DroVir_CAF1 243196 94 + 1 ACACCCGUUCCGCGACCCAGCUC-AACCGAAAUGAACAAAAGUUUUAUUGUGUUGCA-UGAACUCUUAGCAGAAACGUUGAUU-UC---GCAUUCACAA-A- ...........((((..((((.(-((((((...((((....))))..))).))))..-.....(((....)))...))))...-))---))........-.- ( -13.90) >DroSec_CAF1 191519 96 + 1 AUCCCCAGU----CACGCUGUGCGUGCUGGAAUGGUCAACAGUUUUAUUGUCCGACAAAGGACUCUGGAAACAAAUGUUGAUU-UCUGCGCACAGAAAA-AA ....((((.----(((((...)))))))))....((((((((((((...((((......))))....)))))...)))))))(-((((....)))))..-.. ( -32.20) >DroSim_CAF1 177638 96 + 1 AUCCCCAGU----CACGCUGUGCGUGCUGGAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUGGAAACAAAUGUUGAUU-UCUGCGCACAGAAAA-AA ....((((.----(((((...)))))))))....((((((((((((...((((......))))....)))))...)))))))(-((((....)))))..-.. ( -32.20) >DroEre_CAF1 199537 94 + 1 AUCCCCAGG----CACGCAGCGCGUGCAGGAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUAGAAACAAAUGUUGAUU-UCUGCGCACA--AAA-AA ........(----(.....))(.((((((((...(((((((.(((((..((((......))))..))))).....))))))))-))))))).).--...-.. ( -29.30) >DroYak_CAF1 203770 97 + 1 AUUCCCAGU----CACGCUGUGCGUGCUGGAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUAGAAACAAAUGUUGAUUUUCAGCGCACAGAAAA-AA .........----....((((((..(((((((..(((((((.(((((..((((......))))..))))).....))))))))))))))))))))....-.. ( -33.70) >consensus AUCCCCAGU____CACGCUGUGCGUGCUGGAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUGGAAACAAAUGUUGAUU_UCUGCGCACAGAAAA_AA .................((((((((...((...((((((((.(((((..((((......))))..))))).....)))))))).)).))))))))....... (-20.63 = -21.78 + 1.16)


| Location | 2,962,928 – 2,963,030 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Mean single sequence MFE | -22.95 |
| Consensus MFE | -14.38 |
| Energy contribution | -15.17 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
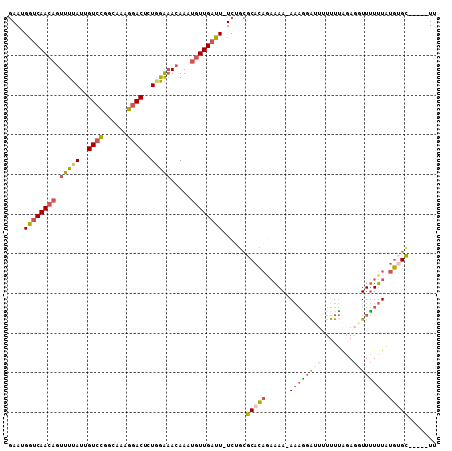
>3L_DroMel_CAF1 2962928 102 + 23771897 GAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUGGAAACAAAUGUUGAUU-UCUGCGCACAGAAAAGAAAAGGAUUUUUUAGAGGUUUUUUAUGUGU-----UU (((..((((((((((((...((((......))))....)))))...))))))))-))...(((((.((((((....((....)).....)))))).)))))-----.. ( -22.90) >DroVir_CAF1 243224 98 + 1 AAAUGAACAAAAGUUUUAUUGUGUUGCA-UGAACUCUUAGCAGAAACGUUGAUU-UC---GCAUUCACAA-A--U-AUU-UUUGGUACAUUAUGUUCAAGUUAUUUUC .((((..(((((((....(((((.(((.-.(((...(((((......))))).)-))---)))..)))))-.--.-)))-))))...))))................. ( -15.20) >DroSec_CAF1 191544 101 + 1 GAAUGGUCAACAGUUUUAUUGUCCGACAAAGGACUCUGGAAACAAAUGUUGAUU-UCUGCGCACAGAAAA-AAAAGAAUGUUUUAGAGGUUUUUUAUGUGC-----UU (((..((((((((((((...((((......))))....)))))...))))))))-)).(((((.((((((-.....((....)).....)))))).)))))-----.. ( -24.70) >DroSim_CAF1 177663 101 + 1 GAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUGGAAACAAAUGUUGAUU-UCUGCGCACAGAAAA-AAAGGAUGUUUUUAGAGGUUUUUUAUGUGC-----UU (((..((((((((((((...((((......))))....)))))...))))))))-))...(((((.....-(((((((.((....)).))))))).)))))-----.. ( -25.80) >DroEre_CAF1 199562 99 + 1 GAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUAGAAACAAAUGUUGAUU-UCUGCGCACA--AAA-AAAGGAUUUUUUUAGAGGUUUUUUAUGAGC-----UU (((..(((((((.(((((..((((......))))..))))).....))))))))-))...((.((--...-((((((((((....)))))))))).)).))-----.. ( -22.00) >DroYak_CAF1 203795 101 + 1 GAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUAGAAACAAAUGUUGAUUUUCAGCGCACAGAAAA-AAAGGAUU-UUUUAGAGGUUUUUUAUGUGC-----UU (((.((((((((.(((((..((((......))))..))))).....)))))))))))((((((.((((((-....((..-..)).....)))))).)))))-----). ( -27.10) >consensus GAAUGGUCAACAGUUUUAUUGUCCGGCAAAGGACUCUGGAAACAAAUGUUGAUU_UCUGCGCACAGAAAA_AAAGGAUUUUUUUAGAGGUUUUUUAUGUGC_____UU ....((((((((.(((((..((((......))))..))))).....))))))))......(((((......((((((((........)))))))).)))))....... (-14.38 = -15.17 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:19 2006