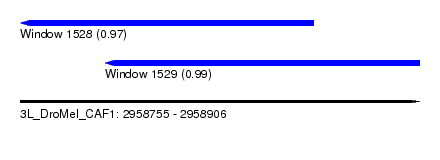
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,958,755 – 2,958,906 |
| Length | 151 |
| Max. P | 0.985270 |

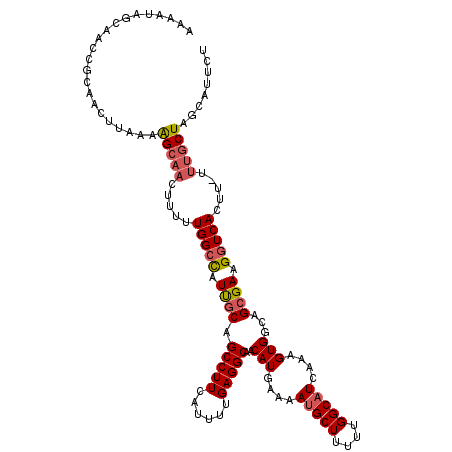
| Location | 2,958,755 – 2,958,866 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.26 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972206 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2958755 111 - 23771897 GCAGCCUUCAUUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUCAAAGUGGCAGCGAAGGUCACUU-UUUGCUAGCAUUCUAGUGUUAUUCAACUAAAACAUUCUAUAUUAAA ...(((((......)))))(((((...((((((....((((..((((((((.......)))))))-).))))))))))...))))).......................... ( -28.50) >DroSec_CAF1 187343 104 - 1 GCAGCCUUCAUUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUCAAAGUGACAGAGAAGGUCACUU-UUUGCUAGCAUUAGAGUGCUAUUCAACUAAAACAUUGUA------- ((((((((......)))))........((((((....((((..((((((((.......)))))))-).))))))))))....)))....................------- ( -27.40) >DroSim_CAF1 173485 111 - 1 GCAGCCUUCAUUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUCAAAGUGACAGCGAAGGUCACUU-UUUGCUAGCAUUCAAGUGCUAUUCAACUAAAACAUUGUAUAUUAAA ((.(((((......))))).((.(((..(((((....((((..((((((((.......)))))))-).)))))))))))).))))........................... ( -27.40) >DroEre_CAF1 195395 104 - 1 GCAGCCUUCAUUUUGAGGCUACAUGCAAAUGCUUUUUGGCUUCAAAGUGGCUGCGAAGGUCA--------CUGGUAAUCUAGUGCUAUUCAACUAAAACAUUGUAUAUUAAA ((((((...((((((((((((...((....))....))))))))))))))))))(((((.((--------((((....)))))))).)))...................... ( -35.90) >DroYak_CAF1 197094 112 - 1 GCAGCCUUCACUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUUUAAGUGGCUGCGAAGGUCAUUUUUUUGCUAGUAUUCUGGUGCUAUUCAACUAAAACACUGUAUAUGAAA ...(((((......)))))..((((.(((((((....))))))).((((((((((((((.....))))))).)))....((((........))))...))))...))))... ( -27.30) >consensus GCAGCCUUCAUUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUCAAAGUGGCAGCGAAGGUCACUU_UUUGCUAGCAUUCUAGUGCUAUUCAACUAAAACAUUGUAUAUUAAA ((((((((......)))))........((((((....((((...(((((((.......)))))))...))))))))))....)))........................... (-21.98 = -22.26 + 0.28)

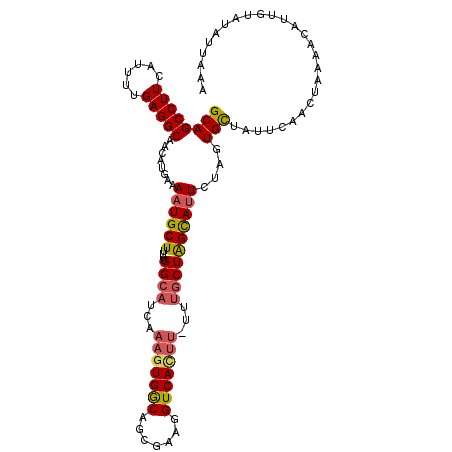
| Location | 2,958,787 – 2,958,906 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.94 |
| Mean single sequence MFE | -34.54 |
| Consensus MFE | -25.28 |
| Energy contribution | -26.00 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2958787 119 - 23771897 AGAAUAGCAACCGCUACUUAAAGGCAACUUUUUGGCCAUUGCAGCCUUCAUUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUCAAAGUGGCAGCGAAGGUCACUU-UUUGCUAGCAUUCU (((((.((....(((((((((((....))))...((((..((.(((((......)))))...((....))))....))))....)))))))((((((((....))-)))))).))))))) ( -37.20) >DroSec_CAF1 187368 119 - 1 AAAAUAGCAACCGCAACUUAAAAGCAACUUUUUGGCCAUUGCAGCCUUCAUUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUCAAAGUGACAGAGAAGGUCACUU-UUUGCUAGCAUUAG ....((((((..((.....(((((((..(((.((((....)).(((((......)))))..)).)))..)))))))..))...((((((((.......)))))))-)))))))....... ( -31.40) >DroSim_CAF1 173517 119 - 1 AAAAUAGCAACCGCAACUUAAAAGCAACUUUUUGGCUAUUGCAGCCUUCAUUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUCAAAGUGACAGCGAAGGUCACUU-UUUGCUAGCAUUCA ....((((((..((.....(((((((..(((.((((....)).(((((......)))))..)).)))..)))))))..))...((((((((.......)))))))-)))))))....... ( -32.60) >DroEre_CAF1 195427 111 - 1 AAAAUAACAACCGCUACUUAAAAGGAUUUUUUUGG-CAUUGCAGCCUUCAUUUUGAGGCUACAUGCAAAUGCUUUUUGGCUUCAAAGUGGCUGCGAAGGUCA--------CUGGUAAUCU .......................(((((((..(((-(.((((((((...((((((((((((...((....))....))))))))))))))))))))..))))--------..)).))))) ( -34.10) >DroYak_CAF1 197126 120 - 1 AAAAUAACAACCGCAACUUAAAAGCAACUUUUUGGCCAUCGCAGCCUUCACUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUUUAAGUGGCUGCGAAGGUCAUUUUUUUGCUAGUAUUCU ......................(((((.....(((((.(((((((((((.....))))...(((..(((((((....)))))))..)))))))))).))))).....)))))........ ( -37.40) >consensus AAAAUAGCAACCGCAACUUAAAAGCAACUUUUUGGCCAUUGCAGCCUUCAUUUUGAGGCAACAUGAAAAUGCUUUUUGGCAUCAAAGUGGCAGCGAAGGUCACUU_UUUGCUAGCAUUCU ......................(((((.....(((((.((((.(((((......)))))..(((....(((((....)))))....)))...)))).))))).....)))))........ (-25.28 = -26.00 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:15 2006