
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 353,762 – 353,863 |
| Length | 101 |
| Max. P | 0.721972 |

| Location | 353,762 – 353,863 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 79.12 |
| Mean single sequence MFE | -17.05 |
| Consensus MFE | -12.68 |
| Energy contribution | -12.48 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.721972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 353762 101 - 23771897 UUUGU-AUU-ACGUAUUACCAUC--CC--AU-CCACUGCUUAUCUACAGGCCAGCGAUGCAACGAGCAAUUGCAACAACGGUAACAAUGCU---AGUAACAAUGCUAAAAA .((((-(((-(.((((((((...--..--..-.....((((......))))..(((((((.....)).)))))......))))...)))))---))).))))......... ( -17.70) >DroVir_CAF1 2852 93 - 1 UCUGUUUAC-ACAAAUUACCAUC--CC--AU-CCACUGCUUAUCUACAGGCCAGCGAUGCAACCAGCAAUUGCA---ACGGUAGCAAUAAC---------AAUGGUAAAAA .........-.....(((((((.--..--..-.(.((((((......))).))).).(((.(((.((....)).---..))).))).....---------.)))))))... ( -17.40) >DroGri_CAF1 145 100 - 1 ------UAU-ACGUAUUACCAUAUCCCAUAU-CCACUGCUUAUCUACAGGCCAGCGAUGCAACGAGCAAUUGCA---ACGGUAACAAUAACAAUACAAACAAUGGUAAAAA ------...-.....(((((((.........-...(((........)))(((.(((((((.....)).))))).---..)))...................)))))))... ( -17.40) >DroWil_CAF1 6823 92 - 1 UAUGU-AUU-ACGUAUUACCAUC--CC--AU-CCACUGCUUAUCUACAGGCCAACGAUGCAACGAGCAAUUGCA---AUGGUAACAA---------UAACAAUGCUAAAAA ...((-(((-.....(((((((.--.(--((-(....((((......))))....))))......((....)).---)))))))...---------....)))))...... ( -14.30) >DroMoj_CAF1 5679 96 - 1 UCUGUUAGCCAUUUUUUACCAUCA-CC--AUCCCAAUGCUUAUCUACAGGCGAGCGAUGCAACCAGCAAUUGCA---GCGGUAGCAAUACA---------AAUGGUAAAAA ............((((((((((..-..--.......(((((..(....)..))))).(((.(((.((.......---))))).))).....---------.)))))))))) ( -22.20) >DroAna_CAF1 8497 92 - 1 UGUGU-AUU-ACGUAUUAUCAUC--CC--AU-CCACUGCUUAUCUACAGGCCAGCGAUGCAUCGAGCAAUUGCA---ACGGUAACAA---------UAACAAUGCUAAAAA ...((-(((-...((((......--..--..-...(((........)))(((.(((((((.....)).))))).---..)))...))---------))..)))))...... ( -13.30) >consensus UCUGU_AUU_ACGUAUUACCAUC__CC__AU_CCACUGCUUAUCUACAGGCCAGCGAUGCAACGAGCAAUUGCA___ACGGUAACAAUA_______UAACAAUGCUAAAAA ...............(((((.................((((......))))..(((((((.....)).)))))......)))))........................... (-12.68 = -12.48 + -0.19)

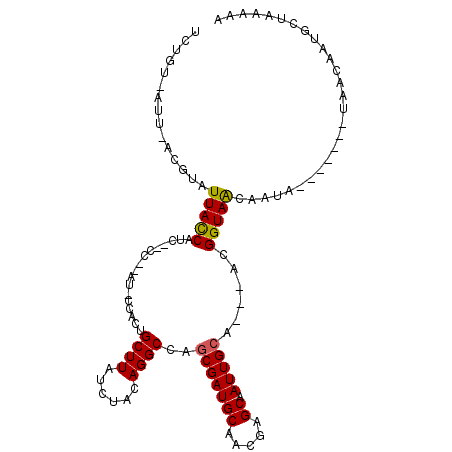
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:19 2006