| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
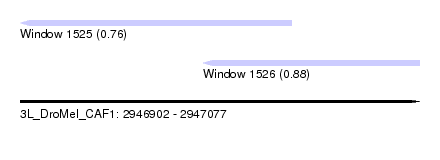
| Location | 2,946,902 – 2,947,077 |
| Length | 175 |
| Max. P | 0.875307 |

| Location | 2,946,902 – 2,947,021 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -22.94 |
| Energy contribution | -23.58 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
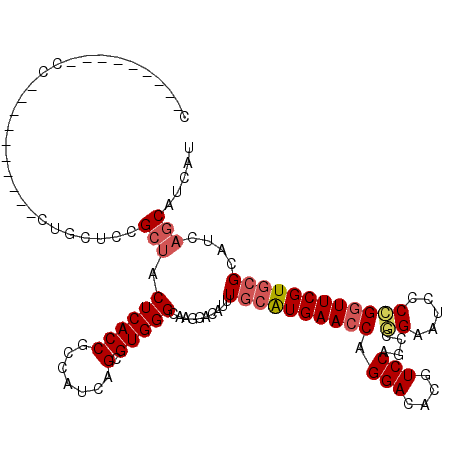
>3L_DroMel_CAF1 2946902 119 - 23771897 CCUGCAGGACCCCUGCUCUGCUCUGCUCCGCUACUCACCGCCAUCAGUGUGGGCAAGGACAUUUGCAUGAACCAGGACACGUCCAGCCUGAAUCCCCGGUUCGUGCGCAUCAGCAUCAU ...(((((...)))))..((((.((((((....((((((.......).)))))...))).....(((((((((.(((....))).....(......)))))))))))))..)))).... ( -38.60) >DroSec_CAF1 182906 100 - 1 C---------CC----------CUGCUCCGCUACUCACCGCCAUCAGCGUGGGCAAGGACAUUUGCAUGAACCAGGACACGUCCAGCCGGAAUCCCCGGUUCGUGCGCAUCAGCAUCAU .---------((----------.(((((((((.............)))).))))).))......(((((((((.(((....)))....((.....)))))))))))((....))..... ( -35.82) >DroSim_CAF1 169049 100 - 1 C---------CC----------CUGCUCCGCUACUCACCGCCAUCAGCGUGGGCAAGGACAUUUGCAUGAACCAGGACACGUCCAGCCGGAAUCCCCGGUUCGUGCGCAUCAGCAUCAU .---------((----------.(((((((((.............)))).))))).))......(((((((((.(((....)))....((.....)))))))))))((....))..... ( -35.82) >DroEre_CAF1 190949 91 - 1 ----------------------------UGCUACUCACCGCCAUUAGAGUGGGCAAGGACAUUUGCAUGAACCAGGACACGUCCAGCCGGAAUCCCCGGUUCGUGCGCAUCAGCAUCAU ----------------------------((((.((((((.......).)))))...........(((((((((.(((....)))....((.....))))))))))).....)))).... ( -29.10) >DroYak_CAF1 192568 114 - 1 CCUUCAGGACCUCUGCA-----CUACUCUGCUACUCACCGCCAUCAGUGUGGGCAAGGACAUUUGCAUGAACCAGGACACGUCCAGCCGGAAUCCCCGGUUCGUGCGCAUCAGCAUCAU ((....)).(((.(((.-----((((.(((..............))).))))))))))......(((((((((.(((....)))....((.....)))))))))))((....))..... ( -35.24) >DroMoj_CAF1 224267 94 - 1 ------------------------AGGGAG-CACUCACCGACAUCAGGGUGGGCAGCGACAUUUUGGUGAACCAGGAUACGUCCAGGCUGAAGCUCAGAUUCGUGCGCAUUAUCAUCAU ------------------------...(((-(.((((((........))))))((((((((((((((....)))))))..)))...))))..))))....................... ( -28.70) >consensus C_________CC__________CUGCUCCGCUACUCACCGCCAUCAGCGUGGGCAAGGACAUUUGCAUGAACCAGGACACGUCCAGCCGGAAUCCCCGGUUCGUGCGCAUCAGCAUCAU .............................(((.((((((.......).)))))..........((((((((((.(((....)))....((.....))))))))))))....)))..... (-22.94 = -23.58 + 0.64)

| Location | 2,946,982 – 2,947,077 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -26.83 |
| Energy contribution | -27.17 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
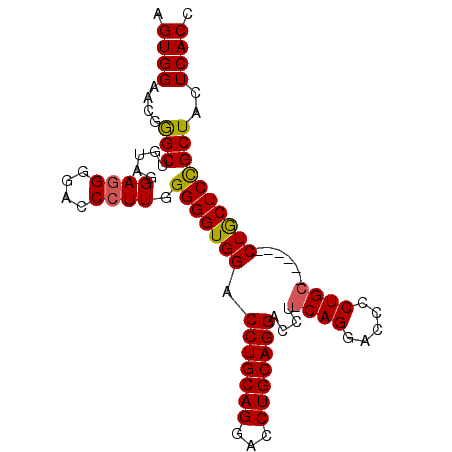
>3L_DroMel_CAF1 2946982 95 - 23771897 AGUGGAACUGGCUGUAGGAGGGGACCCUUGGGGGUGGACCUGCAGGACCUGCAGGACCUGCAGGACCCCUGCUCUGCUCUGCUCCGCUACUCACC ((((((.(.(((.(.((.(((((((((....))))...((((((((.((....)).)))))))).))))).))).)))..).))))))....... ( -48.70) >DroSim_CAF1 169129 76 - 1 AGUGGAACGGGCUGUAGGAGGGGACCCUUGGGGGUGGACCUGCAGGACCUGCAGGAC---------CC----------CUGCUCCGCUACUCACC .((((....(((.(.((.(((((((((....))))...(((((((...))))))).)---------))----------)).))).)))..)))). ( -35.90) >DroYak_CAF1 192648 90 - 1 AGUGGAACGAGCUGUAGGAUGGGAUCCUUUGGGGUGGACCUGCAGGACCUGCAGGACCUUCAGGACCUCUGCA-----CUACUCUGCUACUCACC .((((....(((.((((...(((.((((..(((((...(((((((...)))))))))))).)))).)))....-----))))...)))..)))). ( -32.80) >consensus AGUGGAACGGGCUGUAGGAGGGGACCCUUGGGGGUGGACCUGCAGGACCUGCAGGACCU_CAGGACCCCUGC______CUGCUCCGCUACUCACC .((((....(((.....((((....)))).(((((((.(((((((...)))))))....((((.....))))......))))))))))..)))). (-26.83 = -27.17 + 0.34)

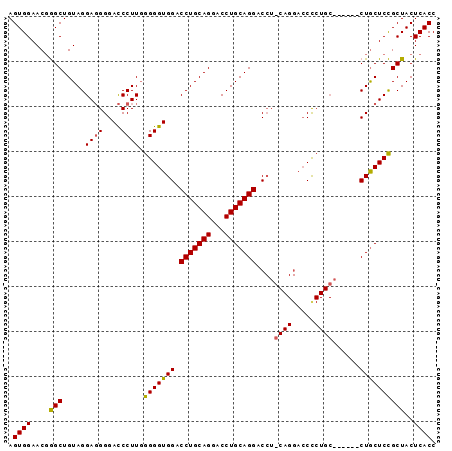
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:12 2006