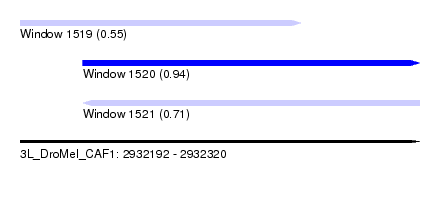
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,932,192 – 2,932,320 |
| Length | 128 |
| Max. P | 0.942874 |

| Location | 2,932,192 – 2,932,282 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.39 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -31.26 |
| Energy contribution | -31.48 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.551138 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2932192 90 + 23771897 GAACGGAAAUGGAGGUGCAACCGGAGG---AGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCAAGUGCGGCAACAGCCGUGGCCACCGC ...(((...((..(((.((.((.....---.))..)))))..))...((((((((((.(((((((...)))))))....))))))))))))). ( -40.80) >DroSec_CAF1 168243 93 + 1 GAACGGAAAUGGAGGAGCAAUCGGAGGAGGAGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGC ...(((...((..((.((..((.......))...))..))..))...((((((((((.(((((((...)))))))....))))))))))))). ( -38.30) >DroSim_CAF1 154422 93 + 1 GAACGGAAAUGGAGGUGCAACCGGAGGAGGAGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGC ...(((......((((....((......)).((.....))....))))(((((((((.(((((((...)))))))....))))))))).))). ( -40.70) >DroEre_CAF1 176131 90 + 1 CAACGGAAAUGGAGGUGCAACCGGAGG---AGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACAGC ....((...((..(((.((.((.....---.))..)))))..)).))((((((((((.(((((((...)))))))....)))))))))).... ( -38.00) >DroYak_CAF1 177499 93 + 1 AAACGGAAAUGGAGGUGCGACCGGAGGAGGAGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACAGC ............((((....((......)).((.....))....))))(((((((((.(((((((...)))))))....)))))))))..... ( -37.10) >DroAna_CAF1 154928 87 + 1 GAG---CAAUGGAACCGGAACCGGAAG---CGGAGUGGCCAACAACCUGGCCAUGGUGGCGGCCUCGAGUGCGGCUACCGCUGUGGCCACCGC ...---........(((....)))..(---(((.((.....))....((((((((((((..(((........)))..)))))))))))))))) ( -39.60) >consensus GAACGGAAAUGGAGGUGCAACCGGAGG___AGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGC .........(((........)))...........(((((((......)))))))(((((((((.....))(((((....)))))))))))).. (-31.26 = -31.48 + 0.22)

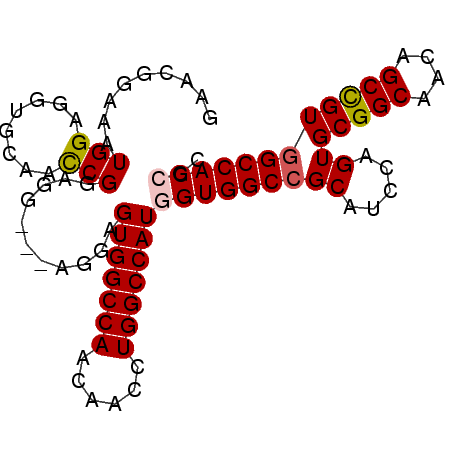
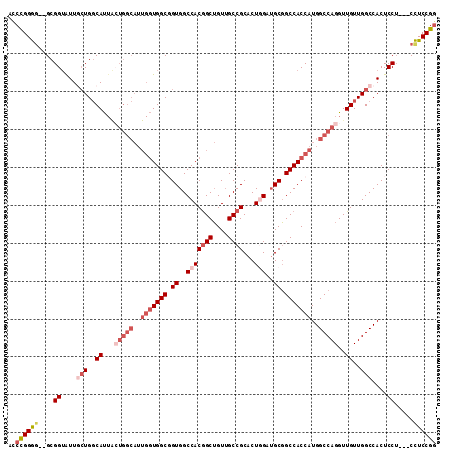
| Location | 2,932,212 – 2,932,320 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -51.49 |
| Consensus MFE | -42.03 |
| Energy contribution | -41.78 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942874 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2932212 108 + 23771897 CCGGAGG---AGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCAAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAGCAAUACCGC--CCCCGGGU ((((.((---.((.(((((........((((((((((.(((((((...)))))))....))))))))))..)))))...(((((....))...)))...)).)--).)))).. ( -52.30) >DroSec_CAF1 168263 111 + 1 UCGGAGGAGGAGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAUCAAUACCGC--CCCCGGGU ((((.((.((.((.(((((((......)))))))(((((((((((...))))(((....)))..)))))))....)).(((.((....)).))).....)).)--).)))).. ( -49.80) >DroSim_CAF1 154442 111 + 1 CCGGAGGAGGAGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAGCAAUACCGC--CCCCGGGU ((((.((.((.((.(((((((......)))))))(((((((((((...))))(((....)))..)))))))....))..(((((....))...)))...)).)--).)))).. ( -52.80) >DroEre_CAF1 176151 108 + 1 CCGGAGG---AGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACAGCCACCAAUGCCACCAAUGCCAGUAACACCGC--CCCCGGGU ((((.((---.((.(((((((......)))))))((((((.((((...))))(((....)))(((((....)))))....)))))).............)).)--).)))).. ( -51.50) >DroYak_CAF1 177519 111 + 1 CCGGAGGAGGAGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACAGCCACCAAUGCCAGUAAUGCCAGUAACACCGC--UCCCGGAA ((((..(.((..(.(((((........((((((((((.(((((((...)))))))....))))))))))..))))))..((.((....)).))......)).)--..)))).. ( -50.30) >DroAna_CAF1 154945 97 + 1 CCGGAAG---CGGAGUGGCCAACAACCUGGCCAUGGUGGCGGCCUCGAGUGCGGCUACCGCUGUGGCCACCGCCA------------GUGGCAGCAACUCCGCAGCUCCAGG- ..(((.(---((((((.((((......((((((((((((..(((........)))..))))))))))))......------------.))))....)))))))...)))...- ( -52.22) >consensus CCGGAGG___AGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAGCAACACCGC__CCCCGGGU ((((.((....((.(((((........((((((((((.(((((((...)))))))....))))))))))..)))))............((....))...))....)))))).. (-42.03 = -41.78 + -0.25)

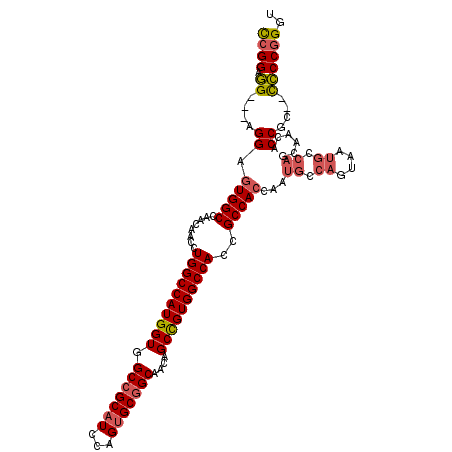
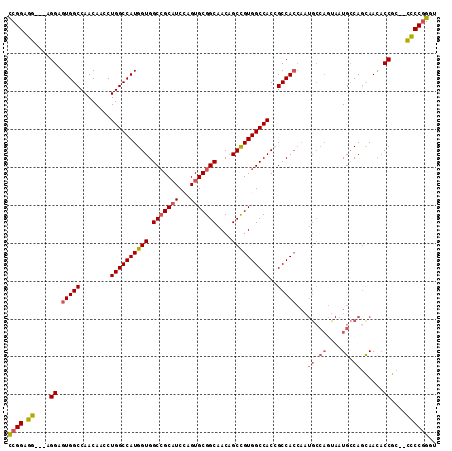
| Location | 2,932,212 – 2,932,320 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.31 |
| Mean single sequence MFE | -57.87 |
| Consensus MFE | -41.14 |
| Energy contribution | -43.58 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2932212 108 - 23771897 ACCCGGGG--GCGGUAUUGCUGGCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUUGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCCU---CCUCCGG ..((((((--(.((....((..(((...(((((..(((((((((..((....))..))..((((.....)))))))))))..)))))..)))..))....)).---))))))) ( -59.60) >DroSec_CAF1 168263 111 - 1 ACCCGGGG--GCGGUAUUGAUGGCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCCUCCUCCUCCGA ...(((((--(.((....(.((((....(((((..(((((((.((..(((((((....))))...)))..)).)))))))..))))).......)))))....)).)))))). ( -57.30) >DroSim_CAF1 154442 111 - 1 ACCCGGGG--GCGGUAUUGCUGGCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCCUCCUCCUCCGG ..((((((--(.((....((..(((...(((((..(((((((.((..(((((((....))))...)))..)).)))))))..)))))..)))..)).......)).))))))) ( -62.00) >DroEre_CAF1 176151 108 - 1 ACCCGGGG--GCGGUGUUACUGGCAUUGGUGGCAUUGGUGGCUGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCCU---CCUCCGG ..((((((--(.((.((..(..(((....((((..((((((((((..(((((((....))))...)))..))))))))))..))))...)))..)..)).)).---))))))) ( -60.40) >DroYak_CAF1 177519 111 - 1 UUCCGGGA--GCGGUGUUACUGGCAUUACUGGCAUUGGUGGCUGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCCUCCUCCUCCGG ..((((((--(.((.((..(..(((...(((((..((((((((((..(((((((....))))...)))..))))))))))..)))))..)))..)..)).))..)))..)))) ( -57.40) >DroAna_CAF1 154945 97 - 1 -CCUGGAGCUGCGGAGUUGCUGCCAC------------UGGCGGUGGCCACAGCGGUAGCCGCACUCGAGGCCGCCACCAUGGCCAGGUUGUUGGCCACUCCG---CUUCCGG -.((((((..(((((((.(((((((.------------(((.(((((((.(.((((...))))....).))))))).))))))).........))).))))))---))))))) ( -50.50) >consensus ACCCGGGG__GCGGUAUUGCUGGCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCCU___CCUCCGG ..((((((....((....(((..((...(((((..(((((((.((..(((((((....))))...)))..)).)))))))..)))))..))..)))....)).....)))))) (-41.14 = -43.58 + 2.45)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:07 2006