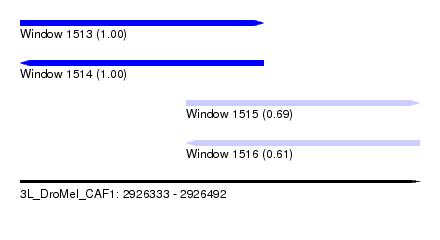
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,926,333 – 2,926,492 |
| Length | 159 |
| Max. P | 0.999934 |

| Location | 2,926,333 – 2,926,430 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 70.28 |
| Mean single sequence MFE | -14.07 |
| Consensus MFE | -8.54 |
| Energy contribution | -8.98 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.61 |
| SVM decision value | 3.81 |
| SVM RNA-class probability | 0.999634 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2926333 97 + 23771897 ACACACGCGCCGAAUACAGCUACACACUAUUGUACCAUAAAUAUUCGCACAC--UCUCUCUCCCUCCU------UCUGGAGUUCUC--------UCUUUUUCUCUUAGUGUAU ..(((((.((.(((((..(.((((......)))).).....))))))).)..--.........((((.------...)))).....--------.............)))).. ( -14.40) >DroEre_CAF1 170224 99 + 1 ACACACGCGCCGACUACAGCUACACACUAUUGUACCAUAAGUAUUCGCACACUCUCUCUCUCCCUCCC------UCUGGAGUUCUC--------UCUUUUUCUCUUAGUGUAU ..(((((((...(((...(.((((......)))).)...)))...)))...............((((.------...)))).....--------.............)))).. ( -15.30) >DroAna_CAF1 149519 102 + 1 ACACACGCGUCAA-UACAAACACAU----AUGUACAA----UUUGCACACAC--UCUCUAUCUCUCUUUUUCUCUUUGGAGUUAUUUCUCCCUUUUUUUUUUUCAUAGUGUAA ..(((((((....-((((.......----.))))...----..)))......--.......................((((......))))................)))).. ( -12.50) >consensus ACACACGCGCCGA_UACAGCUACACACUAUUGUACCAUAA_UAUUCGCACAC__UCUCUCUCCCUCCU______UCUGGAGUUCUC________UCUUUUUCUCUUAGUGUAU ..(((((((...........((((......))))...........)))...............((((..........))))..........................)))).. ( -8.54 = -8.98 + 0.45)

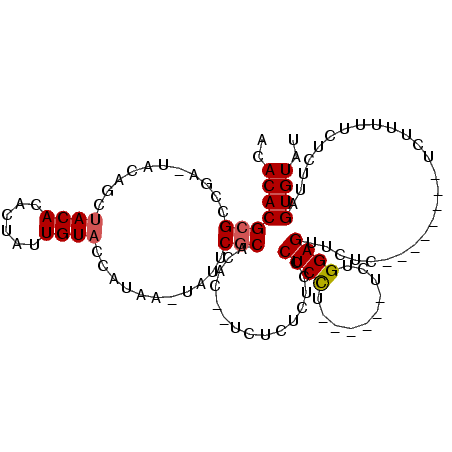
| Location | 2,926,333 – 2,926,430 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 70.28 |
| Mean single sequence MFE | -23.47 |
| Consensus MFE | -13.69 |
| Energy contribution | -14.37 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.58 |
| SVM decision value | 4.65 |
| SVM RNA-class probability | 0.999934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2926333 97 - 23771897 AUACACUAAGAGAAAAAGA--------GAGAACUCCAGA------AGGAGGGAGAGAGA--GUGUGCGAAUAUUUAUGGUACAAUAGUGUGUAGCUGUAUUCGGCGCGUGUGU ((((((.............--------.....((((...------.)))).........--((((.(((((((....(.((((......)))).).))))))))))))))))) ( -25.20) >DroEre_CAF1 170224 99 - 1 AUACACUAAGAGAAAAAGA--------GAGAACUCCAGA------GGGAGGGAGAGAGAGAGUGUGCGAAUACUUAUGGUACAAUAGUGUGUAGCUGUAGUCGGCGCGUGUGU ((((((((...........--------.....((((...------.)))).........((((((....)))))).........)))))))).((((....))))........ ( -22.90) >DroAna_CAF1 149519 102 - 1 UUACACUAUGAAAAAAAAAAAGGGAGAAAUAACUCCAAAGAGAAAAAGAGAGAUAGAGA--GUGUGUGCAAA----UUGUACAU----AUGUGUUUGUA-UUGACGCGUGUGU .(((((................((((......)))).......................--((((((((...----..))))))----))(((((....-..)))))))))). ( -22.30) >consensus AUACACUAAGAGAAAAAGA________GAGAACUCCAGA______AGGAGGGAGAGAGA__GUGUGCGAAUA_UUAUGGUACAAUAGUGUGUAGCUGUA_UCGGCGCGUGUGU .(((((..........................((((..........))))...........((((.(((.(((....(.((((......)))).).))).)))))))))))). (-13.69 = -14.37 + 0.68)

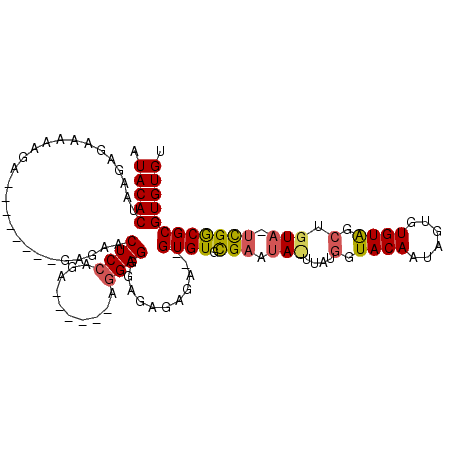
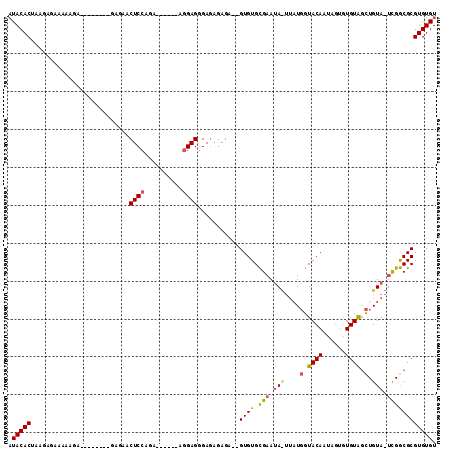
| Location | 2,926,399 – 2,926,492 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -20.05 |
| Consensus MFE | -18.90 |
| Energy contribution | -19.15 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.689237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
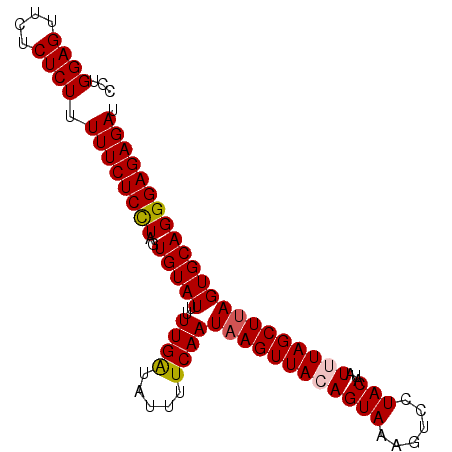
>3L_DroMel_CAF1 2926399 93 + 23771897 UCUGGAGUUCUCUCUUUUUCUCUUAGUGUAUUUUUGGUAUUUUCAAUAAGUUACAGUAAAGUCCUACAGAUUUAGCUUAGUGCAGGGAGAGAU ...((((....))))...((((((..((((((.((((.....))))........(((.(((((.....))))).))).))))))..)))))). ( -18.50) >DroSec_CAF1 162509 93 + 1 CCUGGAGUUCUCUCUUUUUCUCCUAGUGUAUUUUUGAUAUUUUCAAUUAGUUAGAGUAAAGUCCUACAUAUUUAGCUUAGUGCAGGGAGAGAU ...((((....)))).((((((((..((((((.((((.....))))..((((((((((......)))...))))))).)))))))))))))). ( -20.10) >DroSim_CAF1 148650 93 + 1 CCUGGAGUUCUCUCUUUUUCUCCUAGUGUAUUUUUGAUAUUUUCAAUAAGUUAGAGUAAAGUCCUACAUAUUUAGCUUAGUGCAGGGAGAGAU ...((((....)))).((((((((..(((((..((((.....))))((((((((((((......)))...)))))))))))))))))))))). ( -22.60) >DroEre_CAF1 170292 92 + 1 UCUGGAGUUCUCUCUUUUUCUCUUAGUGUAUU-UUGGUGUAUUCAAUAAGUUACAGUAAUUUCAUACAUAUUUAGCUUAGUGCAGGGAGAGAU ........(((((((((........((((((.-(((.((((..........)))).)))....)))))).....((.....))))))))))). ( -19.00) >consensus CCUGGAGUUCUCUCUUUUUCUCCUAGUGUAUUUUUGAUAUUUUCAAUAAGUUACAGUAAAGUCCUACAUAUUUAGCUUAGUGCAGGGAGAGAU ...((((....)))).((((((((..(((((..((((.....))))((((((((((((......)))...)))))))))))))))))))))). (-18.90 = -19.15 + 0.25)

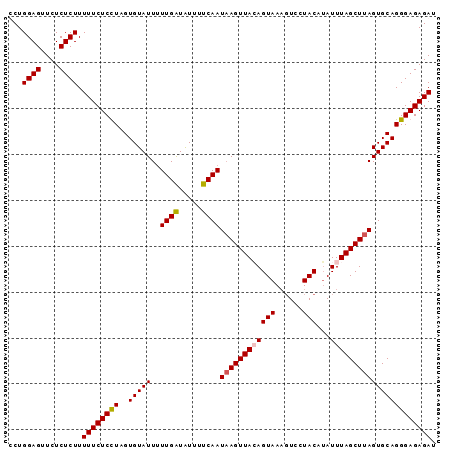
| Location | 2,926,399 – 2,926,492 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -14.85 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.71 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
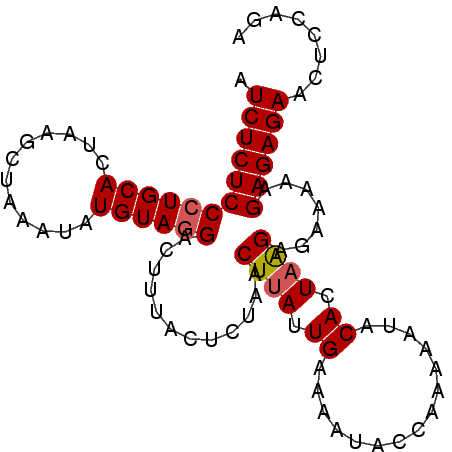
>3L_DroMel_CAF1 2926399 93 - 23771897 AUCUCUCCCUGCACUAAGCUAAAUCUGUAGGACUUUACUGUAACUUAUUGAAAAUACCAAAAAUACACUAAGAGAAAAAGAGAGAACUCCAGA .((((((((((((............)))))).((((..((((..((.(((.......))).))))))...)))).....))))))........ ( -15.80) >DroSec_CAF1 162509 93 - 1 AUCUCUCCCUGCACUAAGCUAAAUAUGUAGGACUUUACUCUAACUAAUUGAAAAUAUCAAAAAUACACUAGGAGAAAAAGAGAGAACUCCAGG .((((((((((((............))))))......((((......((((.....))))..........)))).....))))))........ ( -16.09) >DroSim_CAF1 148650 93 - 1 AUCUCUCCCUGCACUAAGCUAAAUAUGUAGGACUUUACUCUAACUUAUUGAAAAUAUCAAAAAUACACUAGGAGAAAAAGAGAGAACUCCAGG .((((((((((((............))))))......((((....((((............)))).....)))).....))))))........ ( -16.10) >DroEre_CAF1 170292 92 - 1 AUCUCUCCCUGCACUAAGCUAAAUAUGUAUGAAAUUACUGUAACUUAUUGAAUACACCAA-AAUACACUAAGAGAAAAAGAGAGAACUCCAGA .((((((...((.....))......(((((..(((...........)))..)))))....-..................))))))........ ( -11.40) >consensus AUCUCUCCCUGCACUAAGCUAAAUAUGUAGGACUUUACUCUAACUUAUUGAAAAUACCAAAAAUACACUAAGAGAAAAAGAGAGAACUCCAGA .((((((((((((............))))))............((((.((...............)).)))).......))))))........ (-12.46 = -12.71 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:56:03 2006