
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 352,513 – 352,622 |
| Length | 109 |
| Max. P | 0.731193 |

| Location | 352,513 – 352,622 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -31.54 |
| Consensus MFE | -19.69 |
| Energy contribution | -19.72 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.731193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
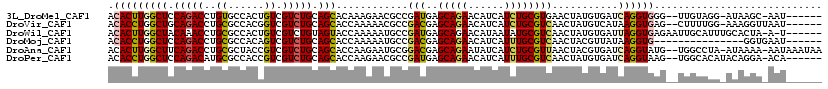
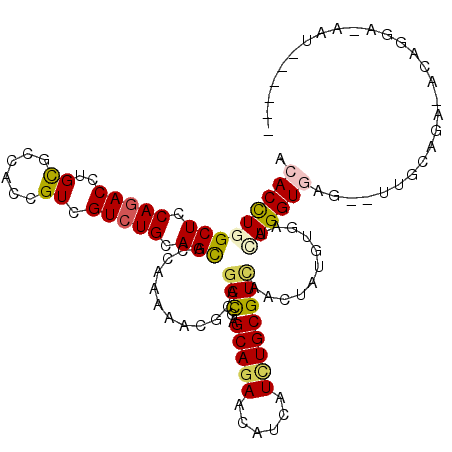
>3L_DroMel_CAF1 352513 109 - 23771897 ACACUUGGCUCCAGACCUGUGCCACUGUCGUCUGCAGCACAAAGAACGCCGAUGAGCAGAACAUCAUCUGCGUGAACUAUGUGAUCAGGUGGG--UUGUAGG-AUAAGC-AAU------ .(((((((((.(((((.............))))).)))(((.((.((((.(((((........))))).))))...)).)))...)))))).(--((((...-....))-)))------ ( -32.12) >DroVir_CAF1 1700 110 - 1 ACACCUGGCUGCAGACCUGCGCCACGGUCGUCUGCAGCACCAAAAACGCCGACGAGCAGAACAUCAUCUGCGUCAACUAUGUCAUAAGGUGAG--CUUUUGG-AAAGGUUAAU------ ..((((.((((((((((((.....)))..))))))))).((((((.((((.....(((((......)))))(.((....)).)....))))..--.))))))-..))))....------ ( -38.20) >DroWil_CAF1 5411 111 - 1 ACACUUGGCUACAAACCUGCGCCACUGUCGUCUGUAGUACCAAAAAUGCCGAUGAGCAGAACAUAAUAUGCGUCAACUAUGUGAUUAGGUGAGAAUUGCAUUUGCACUA-A-U------ .(((((((.((((....(((((..((((((((.(((..........))).))))).)))..........))).))....)))).))))))).....(((....)))...-.-.------ ( -21.60) >DroMoj_CAF1 3912 98 - 1 ACACCUGGCUCCAGACCUGCGCCACAGUCGUCUGCAGCACCAAAAAUGCCGACGAGCAGAACAUCAUUUGCGUCAACUACGUUAUAAGGUG---------------GGUGAAU------ .(((((.(((.((((((((.....)))..))))).))).........((((((..(((((......))))))))(((...)))....))))---------------))))...------ ( -26.40) >DroAna_CAF1 7227 115 - 1 ACACUUGGCUUCAGACCUGCGCUACCGUCGUCUGCAGCACCAAGAAUGCGGACGAGCAGAAUAUCAUCUGCGUUAACUACGUGAUCAGGUAUG--UGGCCUA-AUAAAA-AAUAAAUAA ......((((.((.((((((((.....(((((((((..........)))))))))(((((......))))).........)))..))))).))--.))))..-......-......... ( -37.30) >DroPer_CAF1 9621 110 - 1 ACACCUGGCUCCAGACAUGCGCCACCGUCGUCUGCAGCACCAAGAACGCCGAUGAGCAGAACAUCAUUUGCGUCAACUAUGUGAUCAGGUAAG--UGGCACAUACAGGA-ACA------ ...(((((((.(((((..(((....))).))))).))).....((.(((.(((((........))))).)))))...((((((.(((......--))))))))))))).-...------ ( -33.60) >consensus ACACCUGGCUCCAGACCUGCGCCACCGUCGUCUGCAGCACCAAAAACGCCGACGAGCAGAACAUCAUCUGCGUCAACUAUGUGAUCAGGUGAG__UUGCAGA_ACAGGA_AAU______ .(((((((((.(((((..((......)).))))).)))............(((..(((((......))))))))...........))))))............................ (-19.69 = -19.72 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:18 2006