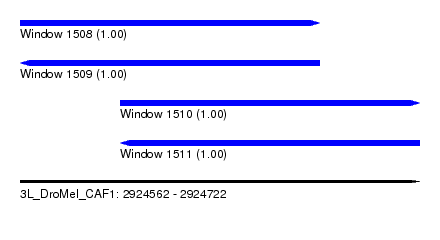
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,924,562 – 2,924,722 |
| Length | 160 |
| Max. P | 0.999916 |

| Location | 2,924,562 – 2,924,682 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -41.67 |
| Energy contribution | -41.53 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.95 |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
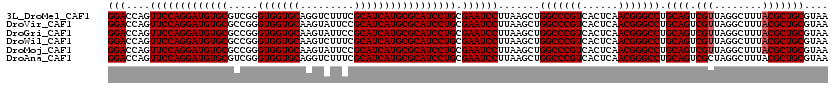
>3L_DroMel_CAF1 2924562 120 + 23771897 UUGUUUCGUUAUUUCUAUUGGAAACGAAUAAGAAUGCAACGGACCAGUUCCAGGAUGUGCGUCGGGUGGUGCAGGUCUUUCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUC .......((...((((.(((....)))...)))).)).((((.(((((((((((((((((.....(((((((.((....))))))))))))))))))).))........))))).)))). ( -42.60) >DroVir_CAF1 204802 120 + 1 UUGUUUCGUUAUUUUUAUUGGAAACGAAUAAGAAUGCAACGGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUAUUCCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUC .......((.((((((((((....).))))))))))).((((.((((((..(((((...(((.(((((.((((.(..........).))))))))).))).)))))..)))))).)))). ( -44.50) >DroGri_CAF1 185029 120 + 1 UUGUUUCGUUAUUUUUAUUGGAAACGAAUAAGAAUGCAACGGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUAUUCCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUC .......((.((((((((((....).))))))))))).((((.((((((..(((((...(((.(((((.((((.(..........).))))))))).))).)))))..)))))).)))). ( -44.50) >DroWil_CAF1 153883 120 + 1 UUGUUUCGCUAUUUCUAUUGGAAACGAAUAAGAAUGCAACGGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUCUUUCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUC .......((.(((..(((((....).))))..))))).((((.((((((..(((((...(((.(((((.((((.(..........).))))))))).))).)))))..)))))).)))). ( -43.90) >DroMoj_CAF1 199013 120 + 1 UUGUUUCGUUAUUUUUAUUGGAAACGAAUAAGAAUGCAACGGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUAUUCCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUC .......((.((((((((((....).))))))))))).((((.((((((..(((((...(((.(((((.((((.(..........).))))))))).))).)))))..)))))).)))). ( -44.50) >DroAna_CAF1 147744 120 + 1 UUGUCUCGUUAUUUCUAUUGGAAACGAAUAAGAAUGCAACGGACCAGUUCCAGGAUGUGCGUCGGGUGGUGCAGGUCUUUCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUC .......((...((((.(((....)))...)))).)).((((.(((((((((((((((((.....(((((((.((....))))))))))))))))))).))........))))).)))). ( -42.60) >consensus UUGUUUCGUUAUUUCUAUUGGAAACGAAUAAGAAUGCAACGGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUAUUCCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUC .......((.((((.(((((....).)))).)))))).((((.(((((((((((((((((.....(((((((.........))))))))))))))))).))........))))).)))). (-41.67 = -41.53 + -0.14)

| Location | 2,924,562 – 2,924,682 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -39.85 |
| Consensus MFE | -38.36 |
| Energy contribution | -38.42 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.37 |
| SVM RNA-class probability | 0.999883 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2924562 120 - 23771897 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACGAAACAA ((((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).))))))))))............(((((...((....))..))))).. ( -38.00) >DroVir_CAF1 204802 120 - 1 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACGAAACAA ((((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))).........((((((............)))))).... ( -40.20) >DroGri_CAF1 185029 120 - 1 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACGAAACAA ((((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))).........((((((............)))))).... ( -40.20) >DroWil_CAF1 153883 120 - 1 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAGCGAAACAA ((((((((((........(((.((((((((((.((.(((((.....))))).))....)))))..))))))))))))))))))............(((((...((....))..))))).. ( -42.40) >DroMoj_CAF1 199013 120 - 1 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACGAAACAA ((((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))).........((((((............)))))).... ( -40.20) >DroAna_CAF1 147744 120 - 1 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACGAGACAA ((((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).))))))))))............(((((...((....))..))))).. ( -38.10) >consensus GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACGAAACAA ((((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))).........((((((............)))))).... (-38.36 = -38.42 + 0.06)
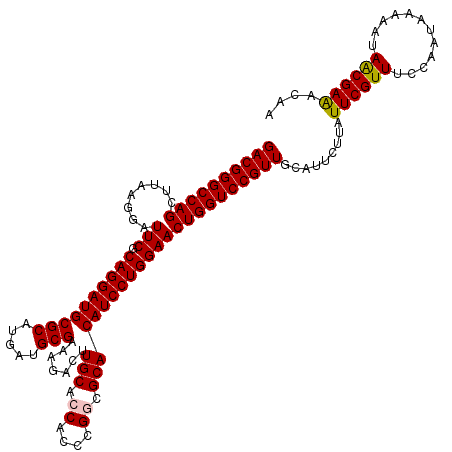
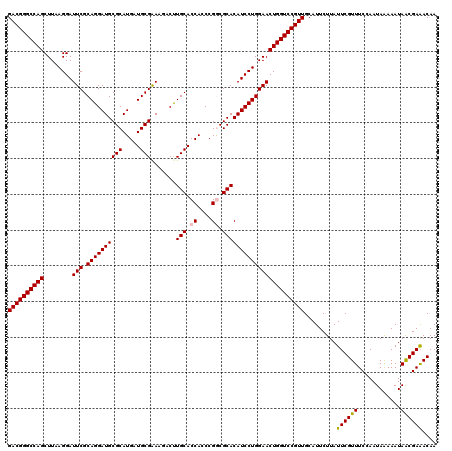
| Location | 2,924,602 – 2,924,722 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -50.78 |
| Consensus MFE | -49.93 |
| Energy contribution | -50.10 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.98 |
| SVM decision value | 4.53 |
| SVM RNA-class probability | 0.999916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2924602 120 + 23771897 GGACCAGUUCCAGGAUGUGCGUCGGGUGGUGCAGGUCUUUCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUCACUCAACGGGCCUGCAGUCGUUAGGCUUUACGCUGCGUAA (((....(((((((((((((.....(((((((.((....))))))))))))))))))).)))))).......(((((((......))))))).((((.(((........))))))).... ( -51.10) >DroVir_CAF1 204842 120 + 1 GGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUAUUCCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUCACUCAACGGGCCUGCAGUCGUUAGGCUUUACGCUGCGUAA (((....(((((((((((((.....(((((((.........))))))))))))))))).)))))).......(((((((......))))))).((((.(((........))))))).... ( -50.60) >DroGri_CAF1 185069 120 + 1 GGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUAUUCCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUCACUCAACGGGCCUGCAGUCGUUAGGCUUUACGCUGCGUAA (((....(((((((((((((.....(((((((.........))))))))))))))))).)))))).......(((((((......))))))).((((.(((........))))))).... ( -50.60) >DroWil_CAF1 153923 120 + 1 GGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUCUUUCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUCACUCAACGGGCCUGCAGUCGUUAGGCUUUACGCUGCGUAA (((....(((((((((((((.....(((((((.........))))))))))))))))).)))))).......(((((((......))))))).((((.(((........))))))).... ( -50.60) >DroMoj_CAF1 199053 120 + 1 GGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUAUUCCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUCACUCAACGGGCCUGCAGUCGUUAGGCUUUACGCUGCGUAA (((....(((((((((((((.....(((((((.........))))))))))))))))).)))))).......(((((((......))))))).((((.(((........))))))).... ( -50.60) >DroAna_CAF1 147784 120 + 1 GGACCAGUUCCAGGAUGUGCGUCGGGUGGUGCAGGUCUUUCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUCACUCAACGGGCCUGCAGUCGCUAGGCUUUACGCUGCGUAA .(((..((((((((((((((.....(((((((.((....))))))))))))))))))).))))......((.(((((((......))))))).)).)))((..(((.....))))).... ( -51.20) >consensus GGACCAGUUCCAGGAUGUGCGCCGGGUGGUGCAAGUAUUCCGCAUCAUGCGCAUCCUGCGAAUCCUUAAGCUGGCCCGUCACUCAACGGGCCUGCAGUCGUUAGGCUUUACGCUGCGUAA (((....(((((((((((((.....(((((((.........))))))))))))))))).)))))).......(((((((......))))))).((((.(((........))))))).... (-49.93 = -50.10 + 0.17)
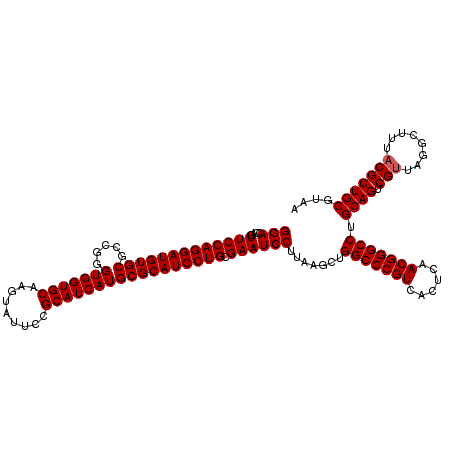
| Location | 2,924,602 – 2,924,722 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -46.42 |
| Consensus MFE | -45.82 |
| Energy contribution | -46.02 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.46 |
| SVM RNA-class probability | 0.999242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2924602 120 - 23771897 UUACGCAGCGUAAAGCCUAACGACUGCAGGCCCGUUGAGUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCC ....(((((((........))).)))).((((((((....)))))))).......((((((.(((((((.((.((.((((.......)))).))......)).)))))))))....)))) ( -43.50) >DroVir_CAF1 204842 120 - 1 UUACGCAGCGUAAAGCCUAACGACUGCAGGCCCGUUGAGUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCC ....(((((((........))).)))).((((((((....)))))))).......((((((.((((((((((.....))).......(((.((....)).))))))))))))....)))) ( -47.70) >DroGri_CAF1 185069 120 - 1 UUACGCAGCGUAAAGCCUAACGACUGCAGGCCCGUUGAGUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCC ....(((((((........))).)))).((((((((....)))))))).......((((((.((((((((((.....))).......(((.((....)).))))))))))))....)))) ( -47.70) >DroWil_CAF1 153923 120 - 1 UUACGCAGCGUAAAGCCUAACGACUGCAGGCCCGUUGAGUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCC ....(((((((........))).)))).((((((((....)))))))).......((((((.((((((((((.((.(((((.....))))).))....)))))..)))))))....)))) ( -48.10) >DroMoj_CAF1 199053 120 - 1 UUACGCAGCGUAAAGCCUAACGACUGCAGGCCCGUUGAGUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCC ....(((((((........))).)))).((((((((....)))))))).......((((((.((((((((((.....))).......(((.((....)).))))))))))))....)))) ( -47.70) >DroAna_CAF1 147784 120 - 1 UUACGCAGCGUAAAGCCUAGCGACUGCAGGCCCGUUGAGUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCC ....(((((((........))).)))).((((((((....)))))))).......((((((.(((((((.((.((.((((.......)))).))......)).)))))))))....)))) ( -43.80) >consensus UUACGCAGCGUAAAGCCUAACGACUGCAGGCCCGUUGAGUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCC ....(((((((........))).)))).((((((((....)))))))).......((((((.((((((((((.....))).......(((.((....)).))))))))))))....)))) (-45.82 = -46.02 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:58 2006