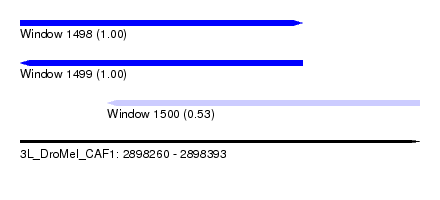
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,898,260 – 2,898,393 |
| Length | 133 |
| Max. P | 0.997518 |

| Location | 2,898,260 – 2,898,354 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -32.65 |
| Consensus MFE | -16.67 |
| Energy contribution | -19.00 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
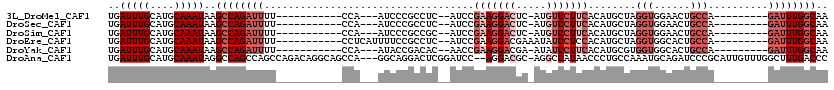
>3L_DroMel_CAF1 2898260 94 + 23771897 UGAUUUGCAUGCAAAUAAGCCAGAUUUU-----------CCA---AUCCCGCCUC--AUCCGAAGGACUC-AUGUCCUUCACAUGCUAGGUGGAACUGCCA---------GAUUUGGCAA ..(((((....)))))..(((((((((.-----------.((---...(((((((--((..(((((((..-..)))))))..)))..))))))...))..)---------)))))))).. ( -35.30) >DroSec_CAF1 142675 94 + 1 UGAUUUGCAUGCAAAUAAGCCAGAUUUU-----------CCA---AUCCCGCCUC--AUCCGAAGGACUC-AUGUCCUUCACAUGCUAGGUGGAACUGCCA---------GAUUUGGCAA ..(((((....)))))..(((((((((.-----------.((---...(((((((--((..(((((((..-..)))))))..)))..))))))...))..)---------)))))))).. ( -35.30) >DroSim_CAF1 133844 94 + 1 UGAUUUGCAUGCAAAUAAGCCAGAUUUU-----------CCA---AUCCCGCCGC--AUCCGAAGGACUC-AUGUCCUUCACAUGCUAGGUGGAACUGCCA---------GAUUUGGCAA ..(((((....)))))..(((((((((.-----------.((---...(((((((--((..(((((((..-..)))))))..))))..)))))...))..)---------)))))))).. ( -37.10) >DroEre_CAF1 149935 98 + 1 UGAUUUGCAUGCAAAUAAGCCAGAUUUU-----------CCUCAUUUUCCGCCUC--AUCCGAAGGACGAAAUAUCCUCCACAUGCUAGGUGGCACUGCCA---------GAUUUGGCAA ..(((((....)))))..(((((((((.-----------...((....(((((((--((..(.((((.......)))).)..)))..))))))...))..)---------)))))))).. ( -24.00) >DroYak_CAF1 151128 94 + 1 UGAUUUGCAUGCAAAUAAGCCAGAUUUU-----------CCA---AUACCGACAC--AACCGAAGGACGA-AUAUCCUUCACAUGCGUGGUGGCACUGCCA---------GAUUUGGCAA ..(((((....)))))..(((((((((.-----------...---..(((.((.(--(...((((((...-...))))))...)).)))))(((...))))---------)))))))).. ( -28.00) >DroAna_CAF1 131172 114 + 1 UGAUUUGCAUGCAAAUAGGCCAGCCAGCCAGACAGGCAGCCA---GGCAGGACUCGGAUCC--AGGACGC-AGGCCAUAACCCUGCCAAAUGCAGAUCCCGCAUUGUUUGGCUUUGACCC ..(((((....))))).((.(((..((((((((((((((((.---....)).)).((((((--(....((-(((.......)))))....))..))))).)).))))))))))))).)). ( -36.20) >consensus UGAUUUGCAUGCAAAUAAGCCAGAUUUU___________CCA___AUCCCGCCUC__AUCCGAAGGACGC_AUGUCCUUCACAUGCUAGGUGGAACUGCCA_________GAUUUGGCAA ..(((((....)))))..((((((((...................................(((((((.....)))))))........(((......)))..........)))))))).. (-16.67 = -19.00 + 2.33)

| Location | 2,898,260 – 2,898,354 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.91 |
| Mean single sequence MFE | -37.17 |
| Consensus MFE | -19.90 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.35 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2898260 94 - 23771897 UUGCCAAAUC---------UGGCAGUUCCACCUAGCAUGUGAAGGACAU-GAGUCCUUCGGAU--GAGGCGGGAU---UGG-----------AAAAUCUGGCUUAUUUGCAUGCAAAUCA ((((((....---------)))))).........(((((((((((((..-..)))))))((((--(((.(.((((---(..-----------..))))).))))))))))))))...... ( -38.50) >DroSec_CAF1 142675 94 - 1 UUGCCAAAUC---------UGGCAGUUCCACCUAGCAUGUGAAGGACAU-GAGUCCUUCGGAU--GAGGCGGGAU---UGG-----------AAAAUCUGGCUUAUUUGCAUGCAAAUCA ((((((....---------)))))).........(((((((((((((..-..)))))))((((--(((.(.((((---(..-----------..))))).))))))))))))))...... ( -38.50) >DroSim_CAF1 133844 94 - 1 UUGCCAAAUC---------UGGCAGUUCCACCUAGCAUGUGAAGGACAU-GAGUCCUUCGGAU--GCGGCGGGAU---UGG-----------AAAAUCUGGCUUAUUUGCAUGCAAAUCA ((((((....---------)))))).........((((.((((((((..-..)))))))).))--))(((.((((---(..-----------..))))).))).(((((....))))).. ( -37.80) >DroEre_CAF1 149935 98 - 1 UUGCCAAAUC---------UGGCAGUGCCACCUAGCAUGUGGAGGAUAUUUCGUCCUUCGGAU--GAGGCGGAAAAUGAGG-----------AAAAUCUGGCUUAUUUGCAUGCAAAUCA ((((((....---------)))))).........(((((((((((((.....)))))))((((--(((.((((........-----------....)))).)))))))))))))...... ( -33.20) >DroYak_CAF1 151128 94 - 1 UUGCCAAAUC---------UGGCAGUGCCACCACGCAUGUGAAGGAUAU-UCGUCCUUCGGUU--GUGUCGGUAU---UGG-----------AAAAUCUGGCUUAUUUGCAUGCAAAUCA .(((((....---------)))))(((....)))(((((((((((((..-..)))))))....--..(((((.((---(..-----------..))))))))......))))))...... ( -32.10) >DroAna_CAF1 131172 114 - 1 GGGUCAAAGCCAAACAAUGCGGGAUCUGCAUUUGGCAGGGUUAUGGCCU-GCGUCCU--GGAUCCGAGUCCUGCC---UGGCUGCCUGUCUGGCUGGCUGGCCUAUUUGCAUGCAAAUCA ((((((..((((..((..(((((....((....((((((..(.(((((.-.......--))..))))..))))))---..))..))))).))..))))))))))(((((....))))).. ( -42.90) >consensus UUGCCAAAUC_________UGGCAGUUCCACCUAGCAUGUGAAGGACAU_GAGUCCUUCGGAU__GAGGCGGGAU___UGG___________AAAAUCUGGCUUAUUUGCAUGCAAAUCA (((((...............))))).........(((((((((((((.....))))))).......((((.((((....................)))).))))....))))))...... (-19.90 = -20.88 + 0.98)
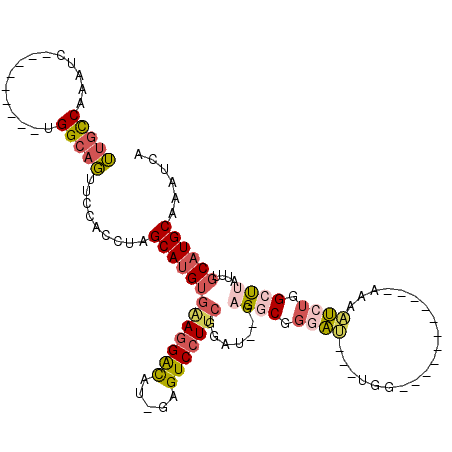
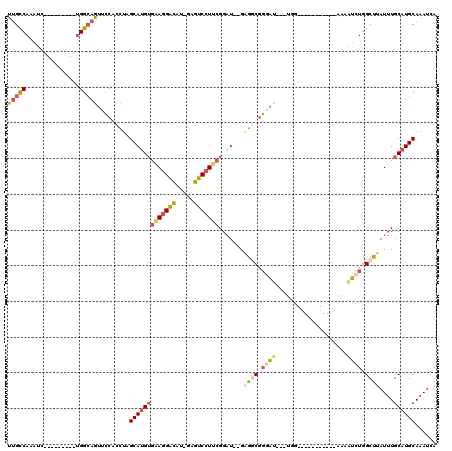
| Location | 2,898,289 – 2,898,393 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.66 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -20.34 |
| Energy contribution | -20.68 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
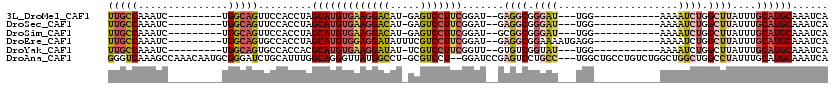
>3L_DroMel_CAF1 2898289 104 - 23771897 -GCGACGAAGCGAUUAACCGUGUCAUUCCCGCAUAUUUGUUUGCCAAAUC---------UGGCAGUUCCACCUAGCAUGUGAAGGACAU-GAGUCCUUCGGAU--GAGGCGGGAU---UG -..(((...(((......))))))..((((((........((((((....---------))))))..........(((.((((((((..-..)))))))).))--)..)))))).---.. ( -38.30) >DroSec_CAF1 142704 104 - 1 -GCGACGAAGCGAUGAACCGUGACAUUCCCGCAUAUUUGCUUGCCAAAUC---------UGGCAGUUCCACCUAGCAUGUGAAGGACAU-GAGUCCUUCGGAU--GAGGCGGGAU---UG -((......))............((.((((((........((((((....---------))))))..........(((.((((((((..-..)))))))).))--)..)))))).---)) ( -37.30) >DroSim_CAF1 133873 104 - 1 -GCGACGAAGCGAUGAACCGUGACAUUCCCGCAUAUUUGCUUGCCAAAUC---------UGGCAGUUCCACCUAGCAUGUGAAGGACAU-GAGUCCUUCGGAU--GCGGCGGGAU---UG -((......))............((.((((((........((((((....---------)))))).........((((.((((((((..-..)))))))).))--)).)))))).---)) ( -41.30) >DroEre_CAF1 149964 108 - 1 -GCGACGAAGCGAUGAGCCGUGACAUUCCCGCAUAUUUGUUUGCCAAAUC---------UGGCAGUGCCACCUAGCAUGUGGAGGAUAUUUCGUCCUUCGGAU--GAGGCGGAAAAUGAG -((......))......((((..(((((((((((......((((((....---------)))))).((......))))))))(((((.....)))))..))))--)..))))........ ( -31.40) >DroYak_CAF1 151157 104 - 1 -GCGACGAAGCGAUGAACCGUGACACUCCCGCAUAUUUGUUUGCCAAAUC---------UGGCAGUGCCACCACGCAUGUGAAGGAUAU-UCGUCCUUCGGUU--GUGUCGGUAU---UG -((......)).....(((..(((((..((((.........(((((....---------)))))(((....)))))....(((((((..-..)))))))))..--))))))))..---.. ( -33.90) >DroAna_CAF1 131212 114 - 1 GGCAGCGAAGCGUUGAACCAUGACAUGCCAGCAUAUUUGCGGGUCAAAGCCAAACAAUGCGGGAUCUGCAUUUGGCAGGGUUAUGGCCU-GCGUCCU--GGAUCCGAGUCCUGCC---UG (((((.((..((..((.(((.((((((....)))....((((((((.((((...((((((((...))))).)))....)))).))))))-))))).)--)).))))..)))))))---.. ( -48.00) >consensus _GCGACGAAGCGAUGAACCGUGACAUUCCCGCAUAUUUGCUUGCCAAAUC_________UGGCAGUUCCACCUAGCAUGUGAAGGACAU_GAGUCCUUCGGAU__GAGGCGGGAU___UG .((......))...................(((....)))(((((...............)))))((((.(((...((.((((((((.....)))))))).))...))).))))...... (-20.34 = -20.68 + 0.34)

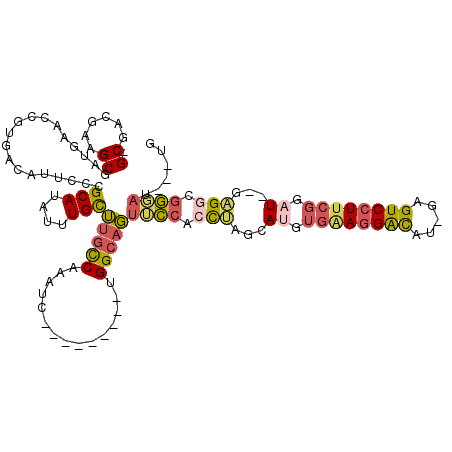
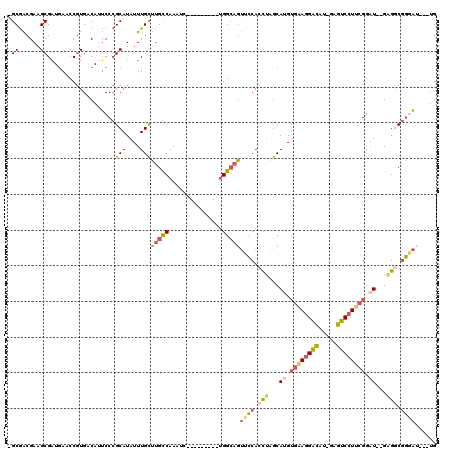
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:47 2006