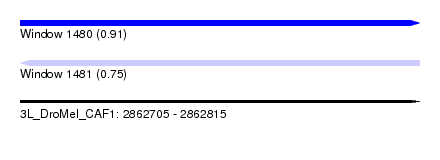
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,862,705 – 2,862,815 |
| Length | 110 |
| Max. P | 0.905559 |

| Location | 2,862,705 – 2,862,815 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 84.34 |
| Mean single sequence MFE | -32.24 |
| Consensus MFE | -21.18 |
| Energy contribution | -22.42 |
| Covariance contribution | 1.24 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
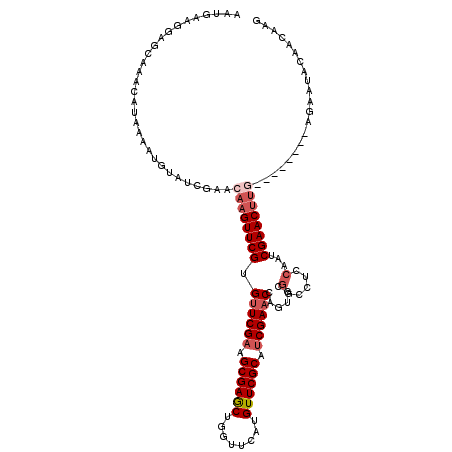
>3L_DroMel_CAF1 2862705 110 + 23771897 AAUGAAGGAGCAAACAUAAAAUGUAUCGAACAAGUUCGUGCUCGAAGCGAACAGGUUCAUGUUCGCAUCGAACCAAUGCGGGCUCCACUCGAACUUA--------AGAAUACAACAAG ......(((((...(((.....(((.((((....)))))))((((.(((((((......))))))).))))....)))...)))))...........--------............. ( -31.60) >DroSec_CAF1 107411 110 + 1 AAUGAAGGAGCAAACAUAAAAUGUAUCGAACAAGUUCGUGUUCGAAGCGAGCUGGUUCAUGUUCGCAUCGAACCAGUGCGGCCUACAAUCGAACUUG--------AGAAUACAACAAG ..((.(((.((..........(((.(((((((......))))))).))).(((((((((((....))).))))))))))..))).))......((((--------.........)))) ( -31.60) >DroSim_CAF1 98112 110 + 1 AAUGAAGGAGCAAACAUAAAAUGUAUCGAACAAGUUCGUGUUCGAAGCGAGCUGGUUCAUGUUCGCAUCGAACCAGUGCGGCCUACAAUCGAACUUG--------AGAAUACAACAAG ..((.(((.((..........(((.(((((((......))))))).))).(((((((((((....))).))))))))))..))).))......((((--------.........)))) ( -31.60) >DroEre_CAF1 111089 118 + 1 AAUGAAGGAGGCAACAUAAAAUGUAUCGAACAAGUUCGCGUUCGCAGCGAGCAGGUUCAUGUUCGCAUCGAACCACCUCGGACGCCAAUCGAACUUGCAUUAAAAAGAACAAAACCAG ......((.(....)....((((((((((....(((((.(((((..(((((((......)))))))..))))).....))))).....))))...)))))).............)).. ( -30.60) >DroYak_CAF1 112799 118 + 1 AAUGAAGGAGGCAACAUAAAAUGUGUCGAACACGUUCGCGUUCGAAGCGAGCUGGUUCAUGUUCGCAUCGAACCAGCACUGCGUCCCAUCGAACUGAAGUGAAAAAGAACCAAACGAG ..(((.(((.(((........(((.((((((.(....).)))))).))).(((((((((((....))).))))))))..))).)))..)))........................... ( -35.80) >consensus AAUGAAGGAGCAAACAUAAAAUGUAUCGAACAAGUUCGUGUUCGAAGCGAGCUGGUUCAUGUUCGCAUCGAACCAGUGCGGCCUCCAAUCGAACUUG________AGAAUACAACAAG ..............................((((((((.((((((.((((((........)))))).))))))......(.....)...))))))))..................... (-21.18 = -22.42 + 1.24)

| Location | 2,862,705 – 2,862,815 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 84.34 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -20.78 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.96 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754391 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
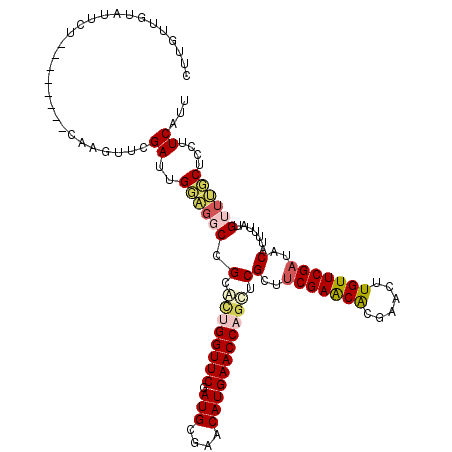
>3L_DroMel_CAF1 2862705 110 - 23771897 CUUGUUGUAUUCU--------UAAGUUCGAGUGGAGCCCGCAUUGGUUCGAUGCGAACAUGAACCUGUUCGCUUCGAGCACGAACUUGUUCGAUACAUUUUAUGUUUGCUCCUUCAUU .....((((((..--------((((((((.((((...))))....((((((.(((((((......))))))).)))))).))))))))...))))))..................... ( -36.80) >DroSec_CAF1 107411 110 - 1 CUUGUUGUAUUCU--------CAAGUUCGAUUGUAGGCCGCACUGGUUCGAUGCGAACAUGAACCAGCUCGCUUCGAACACGAACUUGUUCGAUACAUUUUAUGUUUGCUCCUUCAUU .....((((((..--------((((((((.(((.((((.(..(((((((.(((....))))))))))..))))))))...))))))))...))))))..................... ( -30.60) >DroSim_CAF1 98112 110 - 1 CUUGUUGUAUUCU--------CAAGUUCGAUUGUAGGCCGCACUGGUUCGAUGCGAACAUGAACCAGCUCGCUUCGAACACGAACUUGUUCGAUACAUUUUAUGUUUGCUCCUUCAUU .....((((((..--------((((((((.(((.((((.(..(((((((.(((....))))))))))..))))))))...))))))))...))))))..................... ( -30.60) >DroEre_CAF1 111089 118 - 1 CUGGUUUUGUUCUUUUUAAUGCAAGUUCGAUUGGCGUCCGAGGUGGUUCGAUGCGAACAUGAACCUGCUCGCUGCGAACGCGAACUUGUUCGAUACAUUUUAUGUUGCCUCCUUCAUU ..(((..(((..........(((((((((.((.(((..((((..(((((.(((....))))))))..)))).))).))..)))))))))............)))..)))......... ( -31.55) >DroYak_CAF1 112799 118 - 1 CUCGUUUGGUUCUUUUUCACUUCAGUUCGAUGGGACGCAGUGCUGGUUCGAUGCGAACAUGAACCAGCUCGCUUCGAACGCGAACGUGUUCGACACAUUUUAUGUUGCCUCCUUCAUU .(((.((((............))))..))).((((.((((.((((((((.(((....))))))))))).)(..((((((((....))))))))..).........))).))))..... ( -36.10) >consensus CUUGUUGUAUUCU________CAAGUUCGAUUGGAGGCCGCACUGGUUCGAUGCGAACAUGAACCAGCUCGCUUCGAACACGAACUUGUUCGAUACAUUUUAUGUUUGCUCCUUCAUU ............................((..((((((.(.((((((((.(((....))))))))))).)(..(((((((......)))))))..).......))))))....))... (-20.78 = -21.74 + 0.96)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:29 2006