
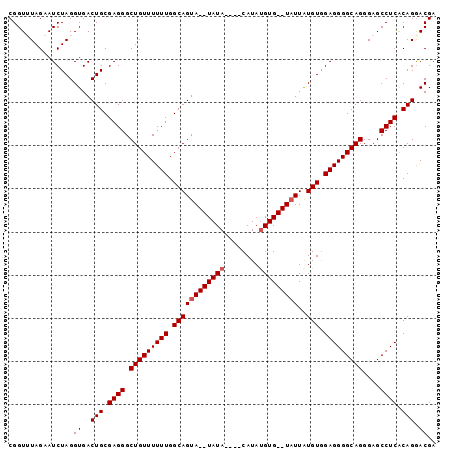
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,858,788 – 2,858,879 |
| Length | 91 |
| Max. P | 0.774562 |

| Location | 2,858,788 – 2,858,879 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -22.06 |
| Energy contribution | -22.86 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.774562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2858788 91 + 23771897 CGGUUUAGAAUCUAGGUGACUGCGAGGGCUGUUUUUUGGCAGUA--UAUA----CAUAUGUG--UAUUAUGUGGAGGGGCAGGGAGCCUCACAGAACGA ..((((.(....(((....))).((((.(((((((((.(((..(--((((----(....)))--)))..))).)))))))))....)))).).)))).. ( -27.30) >DroSec_CAF1 103542 95 + 1 CGGUUUAGAAUCUAGGUGACUGCGAGGGCUGUUUUUUGGCAGUA--UAUACAUACAUAUGUG--UAUUGUGUGGAGGGGCAGGGAGCCUCACAGGACGA ..(((..(....(((....))).((((.(((((((((.(((..(--(((((((....)))))--)))..))).)))))))))....)))).)..))).. ( -29.50) >DroSim_CAF1 94224 95 + 1 CGGUUUAGAAUCUAGGUGACUGCGAGGGCUGUUUUUUGGCAGUA--UAUACAUACAUAUGUG--UAUUAUGUGGAGGGGCAGGGAGCCUCACAGGACGA ..(((..(....(((....))).((((.(((((((((.(((..(--(((((((....)))))--)))..))).)))))))))....)))).)..))).. ( -29.20) >DroEre_CAF1 107216 91 + 1 CGGUAUAGAAUCUAGGUGACUGCGAGGGCUGUUUUUUGGCAGUA--UAUA----CAUACGUG--UAUUAUGUGGAGGGGCAGGGAGCCUCGCAGGACGA ...............((..((((((((.(((((((((.(((..(--((((----(....)))--)))..))).)))))))))....)))))))).)).. ( -32.30) >DroYak_CAF1 108866 95 + 1 CGGUAUAGAAUCUAGGUGACUGCGAGGGCUGUUUUUUGGCAGUACAUAUA----CAUAUGUGUGUUUUAUGUAGAGGGGCAGGGAGCCUCGCAGGACGA ...............((..((((((((.(((((((((.(((..(((((((----....)))))))....))).)))))))))....)))))))).)).. ( -30.80) >consensus CGGUUUAGAAUCUAGGUGACUGCGAGGGCUGUUUUUUGGCAGUA__UAUA____CAUAUGUG__UAUUAUGUGGAGGGGCAGGGAGCCUCACAGGACGA ...............((..(((.((((.(((((((((.((((((((((((........)))))))))..))).)))))))))....)))).))).)).. (-22.06 = -22.86 + 0.80)


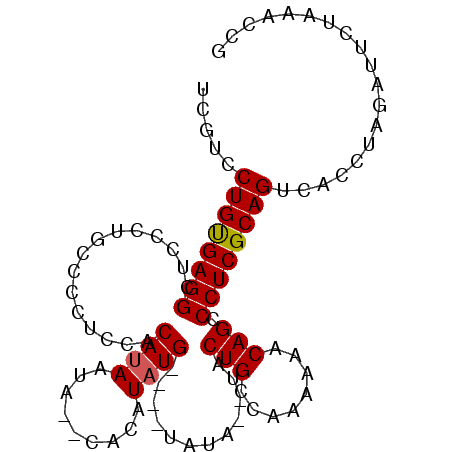
| Location | 2,858,788 – 2,858,879 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 92.45 |
| Mean single sequence MFE | -16.34 |
| Consensus MFE | -12.92 |
| Energy contribution | -12.88 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2858788 91 - 23771897 UCGUUCUGUGAGGCUCCCUGCCCCUCCACAUAAUA--CACAUAUG----UAUA--UACUGCCAAAAAACAGCCCUCGCAGUCACCUAGAUUCUAAACCG ..(..((((((((...................(((--((....))----))).--..(((........))).)))))))).)................. ( -15.10) >DroSec_CAF1 103542 95 - 1 UCGUCCUGUGAGGCUCCCUGCCCCUCCACACAAUA--CACAUAUGUAUGUAUA--UACUGCCAAAAAACAGCCCUCGCAGUCACCUAGAUUCUAAACCG .....((((((((................(((.((--((....)))))))...--..(((........))).))))))))................... ( -15.70) >DroSim_CAF1 94224 95 - 1 UCGUCCUGUGAGGCUCCCUGCCCCUCCACAUAAUA--CACAUAUGUAUGUAUA--UACUGCCAAAAAACAGCCCUCGCAGUCACCUAGAUUCUAAACCG .....((((((((...................(((--((....))))).....--..(((........))).))))))))................... ( -15.20) >DroEre_CAF1 107216 91 - 1 UCGUCCUGCGAGGCUCCCUGCCCCUCCACAUAAUA--CACGUAUG----UAUA--UACUGCCAAAAAACAGCCCUCGCAGUCACCUAGAUUCUAUACCG .....((((((((...................(((--((....))----))).--..(((........))).))))))))................... ( -17.10) >DroYak_CAF1 108866 95 - 1 UCGUCCUGCGAGGCUCCCUGCCCCUCUACAUAAAACACACAUAUG----UAUAUGUACUGCCAAAAAACAGCCCUCGCAGUCACCUAGAUUCUAUACCG .....((((((((.............((((((..(((......))----).))))))(((........))).))))))))................... ( -18.60) >consensus UCGUCCUGUGAGGCUCCCUGCCCCUCCACAUAAUA__CACAUAUG____UAUA__UACUGCCAAAAAACAGCCCUCGCAGUCACCUAGAUUCUAAACCG .....((((((((...............((((.........))))............(((........))).))))))))................... (-12.92 = -12.88 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:26 2006