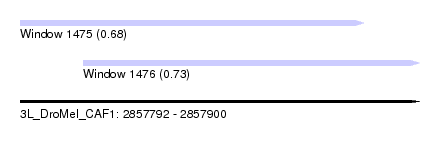
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,857,792 – 2,857,900 |
| Length | 108 |
| Max. P | 0.727996 |

| Location | 2,857,792 – 2,857,885 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.81 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -14.55 |
| Energy contribution | -14.55 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
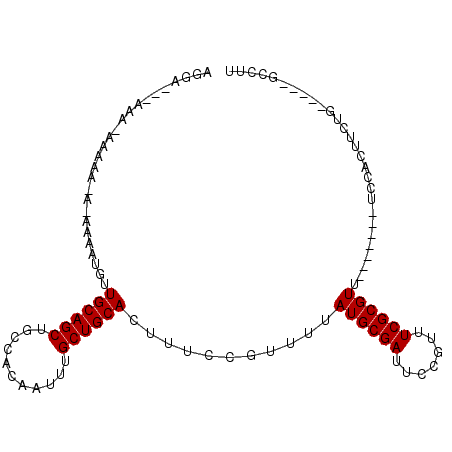
>3L_DroMel_CAF1 2857792 93 + 23771897 --------GAAGAGAACCUGCAGCGAGGAAAA-AAAAACACAAAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU --------...(.(((..(((((((((.....-...((((.....))))((....)).....)))))))))..)))).....((((((.......)))))). ( -21.50) >DroPse_CAF1 122210 75 + 1 --------GGAGAGAACCUGCAG-------------------AAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU --------((((((.....(((.-------------------...)))((((((...........)))))))))))).....((((((.......)))))). ( -25.00) >DroSec_CAF1 102532 94 + 1 --------GAAGAGAACCUGCAGCGAGGAAAAAAAAAACACAAAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU --------...(.(((..(((((((((.........((((.....))))((....)).....)))))))))..)))).....((((((.......)))))). ( -21.50) >DroMoj_CAF1 122066 86 + 1 --------AAA--GA--GCA-GAAGAGAGAACAAAAAA---UUGAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGACUGCGUUUCGCGUU --------...--..--(((-(..(((((..(((....---)))....((((((...........)))))))))))(((.....))).)))).......... ( -20.40) >DroAna_CAF1 91842 94 + 1 CUAAAACUGUAGAGAACCUGCAGCA--GA------AGAGAAAAAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU .((((((.(.((((....(((((((--((------..........(((.((....)).))).)))))))))))))).))))))(((((.......))))).. ( -22.60) >DroPer_CAF1 123161 75 + 1 --------GGAGAGAACCUGCAG-------------------AAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU --------((((((.....(((.-------------------...)))((((((...........)))))))))))).....((((((.......)))))). ( -25.00) >consensus ________GAAGAGAACCUGCAGCG__GA______AAA___AAAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU ................................................((((((...........))))))...........((((((.......)))))). (-14.55 = -14.55 + -0.00)


| Location | 2,857,809 – 2,857,900 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -14.40 |
| Energy contribution | -14.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.727996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2857809 91 + 23771897 AGGA---AAA-AAAAACACAAAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU-------UCCACUUCUG-----GCCUU .(((---((.-........((((((.((((((...........)))))).....)))))).(((((.......)))))))-------))).......-----..... ( -20.30) >DroGri_CAF1 122343 89 + 1 AGAA---GGAAAAAAA---UUGAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGACUUCGUUUCGCGUU-------UCCACUUUGGC-----CAGC ....---(((((((((---(.((...((((((...........))))))...)).))))).(((((.......)))))))-------)))........-----.... ( -21.30) >DroWil_CAF1 94221 82 + 1 AGGA---AGA-------AAAAACUUUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU-------UCCACUUUUU--------AU .(((---(..-------.(((((...((((((...........))))))......)))))((((((.......)))))))-------))).......--------.. ( -19.70) >DroYak_CAF1 107845 94 + 1 AGGAAGAAAA-AAAAACACAAAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU-------UCCACUUCUG-----GCCUU (((.((((..-..((((..((((((.((((((...........)))))).....))))))..((((.......)))))))-------)....)))).-----.))). ( -21.80) >DroMoj_CAF1 122078 101 + 1 AGAG---AACAAAAAA---UUGAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGACUGCGUUUCGCGUUUCGCGAUUCCACUUUGGCAUCGGCAGA ....---..((.((((---(.((...((((((...........))))))...)).))))).))...((((.....(((...)))(((.((.....)).))).)))). ( -24.80) >DroAna_CAF1 91867 84 + 1 --GA---------AGAGAAAAAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU-------UCCACUUCUG-----GCCUU --((---------((.(((((((((.((((((...........)))))).....))))))((((((.......)))))))-------))..))))..-----..... ( -19.50) >consensus AGGA___AAA_AAAAA_A_AAAAUGUUGCAGCUGCCACAAUUUGCUGCACUUUCCGUUUUAUGCGAUUCCGUUUCGCGUU_______UCCACUUCUG_____GCCUU ..........................((((((...........))))))...........((((((.......))))))............................ (-14.40 = -14.40 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:25 2006