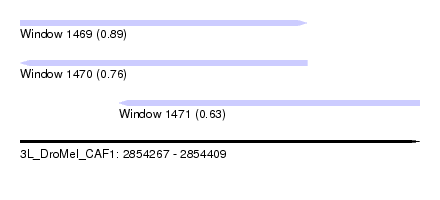
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,854,267 – 2,854,409 |
| Length | 142 |
| Max. P | 0.892262 |

| Location | 2,854,267 – 2,854,369 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -36.95 |
| Energy contribution | -36.73 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
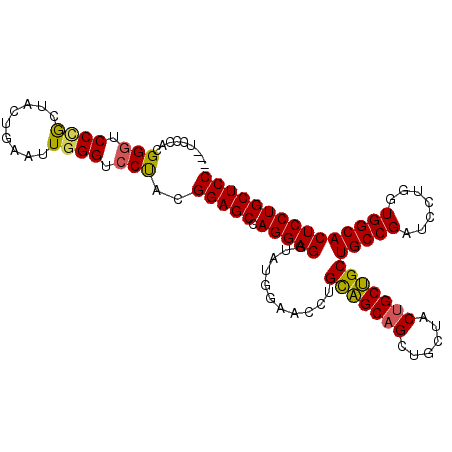
>3L_DroMel_CAF1 2854267 102 + 23771897 ---UCCCACGGGUCCUGCUACUGAAUUGGGUCCUACGGAGCCAGGAGGUAUGGAACCUGUUGCAGCUGCUACUGCAGCUGCCGAUCCUGGUGGCACUCCUGCUCC ---......((..((............))..))...(((((.(((((((.....(((.(((((((((((....))))))).))))...))).)).)))))))))) ( -44.10) >DroSec_CAF1 99074 102 + 1 ---UCCCACGGGUCCCACUACUGAAUUGGGACCUACGGAGCCAGGAGGUAUGGAACCUGCAGCAGCUGCUACUGCUGCUGCCGAUCCUGGUGGCACUCCUGCUCC ---......((((((((.........))))))))..(((((.(((((((..(((.(..((((((((.......)))))))).).))).....)).)))))))))) ( -49.20) >DroYak_CAF1 104183 105 + 1 AGAUCCCACGGAUCCCGUUACUGAGUUUGGUCCCACGGAGCCAGGAGGUAGGACACCUGCAGCAGCUGCGACCGCUGCUGCCGAUCCAGGUGGCACUCCUGCUCC .(((((...)))))..(..(((......)))..)..(((((.(((((....(.(((((((((((((.......))))))))......))))).).)))))))))) ( -42.60) >consensus ___UCCCACGGGUCCCGCUACUGAAUUGGGUCCUACGGAGCCAGGAGGUAUGGAACCUGCAGCAGCUGCUACUGCUGCUGCCGAUCCUGGUGGCACUCCUGCUCC .........(((.((((.........)))).)))..(((((.(((((...........(((((((......)))))))(((((.......))))))))))))))) (-36.95 = -36.73 + -0.21)

| Location | 2,854,267 – 2,854,369 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 87.18 |
| Mean single sequence MFE | -45.40 |
| Consensus MFE | -37.40 |
| Energy contribution | -37.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761281 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2854267 102 - 23771897 GGAGCAGGAGUGCCACCAGGAUCGGCAGCUGCAGUAGCAGCUGCAACAGGUUCCAUACCUCCUGGCUCCGUAGGACCCAAUUCAGUAGCAGGACCCGUGGGA--- (((((.((....))..(((((...((((((((....))))))))....(((.....)))))))))))))......((((..((........))....)))).--- ( -41.50) >DroSec_CAF1 99074 102 - 1 GGAGCAGGAGUGCCACCAGGAUCGGCAGCAGCAGUAGCAGCUGCUGCAGGUUCCAUACCUCCUGGCUCCGUAGGUCCCAAUUCAGUAGUGGGACCCGUGGGA--- ((((((((((.(......(((((.((((((((.......)))))))).)))))....)))))).)))))...(((((((.........))))))).......--- ( -48.10) >DroYak_CAF1 104183 105 - 1 GGAGCAGGAGUGCCACCUGGAUCGGCAGCAGCGGUCGCAGCUGCUGCAGGUGUCCUACCUCCUGGCUCCGUGGGACCAAACUCAGUAACGGGAUCCGUGGGAUCU ((((((((((.(.((((((...(((((((.((....)).))))))))))))).)....))))).))))).((((......))))......((((((...)))))) ( -46.60) >consensus GGAGCAGGAGUGCCACCAGGAUCGGCAGCAGCAGUAGCAGCUGCUGCAGGUUCCAUACCUCCUGGCUCCGUAGGACCCAAUUCAGUAGCGGGACCCGUGGGA___ ((((((((((((((.........))))((((((((....))))))))...........))))).)))))......((((..((........))....)))).... (-37.40 = -37.40 + 0.00)

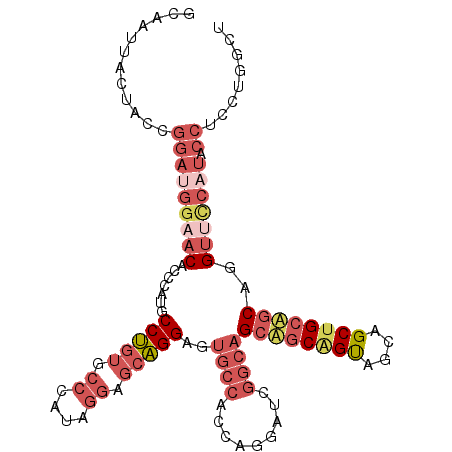
| Location | 2,854,302 – 2,854,409 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 71.34 |
| Mean single sequence MFE | -42.67 |
| Consensus MFE | -18.71 |
| Energy contribution | -23.15 |
| Covariance contribution | 4.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629217 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
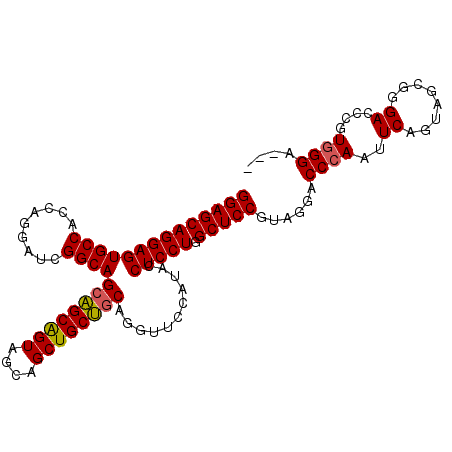
>3L_DroMel_CAF1 2854302 107 - 23771897 GCAAUUACUACCGGAUGGAACACCAAUGCCUGUGCCCAUAGGAGCAGGAGUGCCACCAGGAUCGGCAGCUGCAGUAGCAGCUGCAACAGGUUCCAUACCUCCUGGCU (((............(((....)))...(((((.((....)).)))))..)))..((((((...((((((((....))))))))....(((.....))))))))).. ( -40.60) >DroSec_CAF1 99109 107 - 1 GCAAUUACUACCGGAUGGAACACCCAUGCCUGUGCCCAUAGGAGCAGGAGUGCCACCAGGAUCGGCAGCAGCAGUAGCAGCUGCUGCAGGUUCCAUACCUCCUGGCU (((...........((((.....)))).(((((.((....)).)))))..)))..((((((...((((((((.......)))))))).(((.....))))))))).. ( -41.80) >DroYak_CAF1 104221 107 - 1 GCAAUUACUACCGGAUGGAACACCCAUGCCUGUGCCCAUAGGAGCAGGAGUGCCACCUGGAUCGGCAGCAGCGGUCGCAGCUGCUGCAGGUGUCCUACCUCCUGGCU .........((.((((((.....)))).)).))(((...(((((.((((....((((((...(((((((.((....)).)))))))))))))))))..)))))))). ( -43.10) >DroMoj_CAF1 115913 83 - 1 UCAGCUCAUGCCCGACGGCACGCCCAUGCCGCUGCCGAU------UGGAGCC---GCUGCAGCAGCAGCAGCGGCCGCCGCAGGAGCUGGG---------------C (((((((.((((((((((((.((.......))))))).)------))).(((---(((((.......))))))))....))).))))))).---------------. ( -45.20) >consensus GCAAUUACUACCGGAUGGAACACCCAUGCCUGUGCCCAUAGGAGCAGGAGUGCCACCAGGAUCGGCAGCAGCAGUAGCAGCUGCAGCAGGUUCCAUACCUCCUGGCU ............(((((((((.......(((((.((....)).)))))..((((.........))))((((((((....))))))))..))))))).))........ (-18.71 = -23.15 + 4.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:20 2006