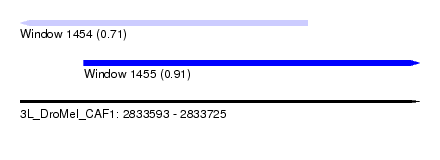
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,833,593 – 2,833,725 |
| Length | 132 |
| Max. P | 0.909303 |

| Location | 2,833,593 – 2,833,688 |
|---|---|
| Length | 95 |
| Sequences | 3 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 93.59 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -23.69 |
| Energy contribution | -24.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
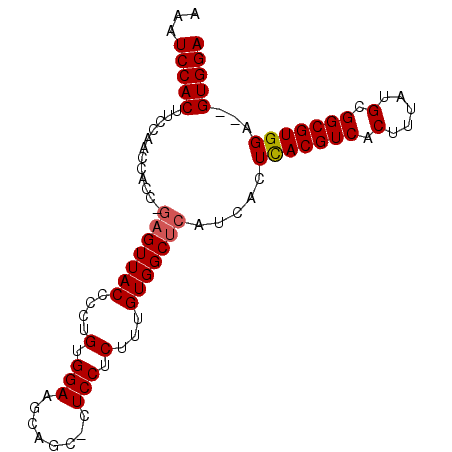
>3L_DroMel_CAF1 2833593 95 - 23771897 AAAUCCACUUCCAACCACCCCUGUUACCCCUGUGGAAGCAGCUCUCCUCUUUGUGGCUCAUCACUCACGUCACUUUAUGCGGCGUGGAGAGUGGA ...(((.((((((...................))))))..(((((((.....((((....))))..(((((.(.....).))))))))))))))) ( -26.71) >DroSec_CAF1 78627 91 - 1 AAAUCCACUUCCAACCACC-GAGUUACCCCUGUGGAAGCAGC-CUCCUCUUUGUGGCUCAUCACUCACGUCACUUUAUGUGGCGUGGA--GUGGA ...(((((((.........-(((((((....(.(((......-.))).)...)))))))......((((((((.....))))))))))--))))) ( -32.70) >DroSim_CAF1 69211 91 - 1 AAAUCCACAUCCAACCACC-GAGUUACCCCUGUGGAAGCAGC-CUCCUCUUUGUGGCUCAUCACUUACGUCACUUUAUGCGGCGUGGA--GUGGA ...(((((.(((.......-(((((((....(.(((......-.))).)...))))))).......(((((.(.....).))))))))--))))) ( -28.00) >consensus AAAUCCACUUCCAACCACC_GAGUUACCCCUGUGGAAGCAGC_CUCCUCUUUGUGGCUCAUCACUCACGUCACUUUAUGCGGCGUGGA__GUGGA ...(((((............(((((((....(.(((........))).)...))))))).....(((((((.(.....).)))))))...))))) (-23.69 = -24.13 + 0.45)

| Location | 2,833,614 – 2,833,725 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 91.57 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -24.64 |
| Energy contribution | -24.48 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909303 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
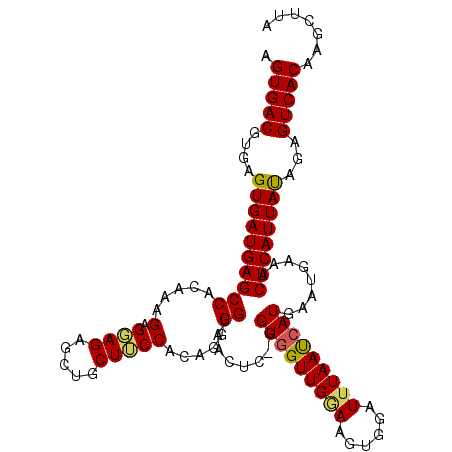
>3L_DroMel_CAF1 2833614 111 + 23771897 AGUGACGUGAGUGAUGAGCCACAAAGAGGAGAGCUGCUUCCACAGGGGUAACAGGGGUGGUUGGAAGUGGAUUUAAUCAUGAAUGAAACUCAUUAUAGAGUCACAAGCUUA .(((((....((((((((((((...(((.(....).)))((((...(....)....))))......))))......(((....)))..))))))))...)))))....... ( -26.70) >DroSec_CAF1 78646 109 + 1 AGUGACGUGAGUGAUGAGCCACAAAGAGGAG-GCUGCUUCCACAGGGGUAACUC-GGUGGUUGGAAGUGGAUUUAAUCAUGAAUGAAACUCAUUACAGAGUCACAAGCUUA .(((((....((((((((((.......))..-.(..(((((((.(((....)))-....).))))))..)..................))))))))...)))))....... ( -32.10) >DroSim_CAF1 69230 109 + 1 AGUGACGUAAGUGAUGAGCCACAAAGAGGAG-GCUGCUUCCACAGGGGUAACUC-GGUGGUUGGAUGUGGAUUUAAUCAUGAAUGAAACUCAUUAUAGAGUCACAAGCUUA .(((((....((((((((..........(((-..((((((....)))))).)))-.((((((((((....))))))))))........))))))))...)))))....... ( -30.90) >DroEre_CAF1 81383 110 + 1 AGUGACGUGAGUGAUGAGCCAGAAGGAGGAGAGCUGCUGCCACACGGGUAACUC-GGUGGUUGGAAGAAGAUGUAAUCAUGAAUGAAACUCAUUACGGAGUCACAAGCUUA .(((((....(((((((((((..((.((.....)).))(((((.((((...)))-)))))))))............(((....)))..))))))))...)))))....... ( -32.40) >DroYak_CAF1 81855 110 + 1 AGUGACGUGAGUGAUGAGCCAGAAAGAGCAGAGCUGCUGCCACACGGGUAACUC-GGUGGUUGAAAGGAGAUUUAACAAUGAAUGAAACUCAUUAUGGAGUCACAAGCUUA .(((((....((((((((((......((((....))))(((((.((((...)))-)))))).....))..(((((....)))))....))))))))...)))))....... ( -32.30) >consensus AGUGACGUGAGUGAUGAGCCACAAAGAGGAGAGCUGCUUCCACAGGGGUAACUC_GGUGGUUGGAAGUGGAUUUAAUCAUGAAUGAAACUCAUUAUAGAGUCACAAGCUUA .(((((....((((((((((.....(.((((.....))))).....))........(((((((((......)))))))))........))))))))...)))))....... (-24.64 = -24.48 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:05 2006