
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,829,383 – 2,829,483 |
| Length | 100 |
| Max. P | 0.848460 |

| Location | 2,829,383 – 2,829,483 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -12.80 |
| Energy contribution | -12.47 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
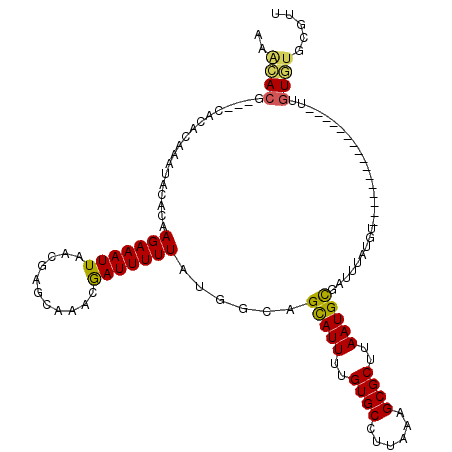
>3L_DroMel_CAF1 2829383 100 + 23771897 AAGCACG---CACACAAAUACACAAGAAAUUAACGAGCAAACGAUUUUUAUGGCAGUAUUUUGUGCCUUAAAGCGCUUAAUGCGAUUUAUGU----------------UUGUGUGCGUU ....(((---((((((((((..(((((((((...........))))))).))...(((((..((((......))))..)))))......)))----------------)))))))))). ( -29.30) >DroGri_CAF1 89154 97 + 1 AAAA------AAAACAAGUACACAAGAAAUUAACGAGCAAACAAUUUUUAUGGCAGCAUUUUGUGCCUUAAAGCGCUUAAUGCGCUUUAUGG------CUAUAUAU----------AGA ....------...(((((....(((((((((...........))))))).)).......)))))(((.((((((((.....)))))))).))------).......----------... ( -19.50) >DroSec_CAF1 74398 100 + 1 AAGCACG---CACACAAAUACACAAGAAAUUAACGAGCAAACGAUUUUUAUUGCAGUAUUUUGUGCCUUAAAGCGCUUAAUGCGAUUUAUGU----------------UUGUGUGCGUU ....(((---((((((((((................((((..........)))).(((((..((((......))))..)))))......)))----------------)))))))))). ( -30.60) >DroWil_CAF1 66836 119 + 1 AAAUACAAAAAACACAAGUAUACAAGAAAUUAACGAGCAGACGAUUUUUAUGGCAGCAUUUUGUGCCUUAAAGCGCUUAAUGUGUUUUAUACAUACAUACAUGCAUAUUAAUAUGAGAA ................(((((.(((((((((..(.....)..))))))).))...((((..((((...((((((((.....))))))))....))))...))))))))).......... ( -16.30) >DroYak_CAF1 77392 100 + 1 AAGCACG---CACACAAAUACACAAGAAAUUAACGAGCAAACGAUUUUUAUGGCAGUAUUUUGUGCCUUAAAGCGCUUAAUGCGAUUUAUGU----------------UUGUGUGCGUU ....(((---((((((((((..(((((((((...........))))))).))...(((((..((((......))))..)))))......)))----------------)))))))))). ( -29.30) >DroAna_CAF1 62780 98 + 1 AAACACG---CACACAAGUACACAAGAAAUUAACGAGCAAACGAUUUUUAUGGCAGCAUUUUGUGCCUUAAAGCGCUUAAUGCGAUUUAUGU----------------GAGUGUG--UU .((((((---(.((((......(((((((((...........))))))).))...(((((..((((......))))..)))))......)))----------------).)))))--)) ( -25.90) >consensus AAACACG___CACACAAAUACACAAGAAAUUAACGAGCAAACGAUUUUUAUGGCAGCAUUUUGUGCCUUAAAGCGCUUAAUGCGAUUUAUGU________________UUGUGUGCGUU ..((((..................(((((((...........)))))))......(((((..((((......))))..)))))...........................))))..... (-12.80 = -12.47 + -0.33)

| Location | 2,829,383 – 2,829,483 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 79.77 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -10.49 |
| Energy contribution | -10.82 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.43 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2829383 100 - 23771897 AACGCACACAA----------------ACAUAAAUCGCAUUAAGCGCUUUAAGGCACAAAAUACUGCCAUAAAAAUCGUUUGCUCGUUAAUUUCUUGUGUAUUUGUGUG---CGUGCUU .((((((((((----------------((((((..(((.....))).((((.((((........)))).)))).....................))))))..)))))))---))).... ( -27.70) >DroGri_CAF1 89154 97 - 1 UCU----------AUAUAUAG------CCAUAAAGCGCAUUAAGCGCUUUAAGGCACAAAAUGCUGCCAUAAAAAUUGUUUGCUCGUUAAUUUCUUGUGUACUUGUUUU------UUUU ...----------.......(------((.((((((((.....)))))))).)))............(((((((((((.........)))))).)))))..........------.... ( -20.70) >DroSec_CAF1 74398 100 - 1 AACGCACACAA----------------ACAUAAAUCGCAUUAAGCGCUUUAAGGCACAAAAUACUGCAAUAAAAAUCGUUUGCUCGUUAAUUUCUUGUGUAUUUGUGUG---CGUGCUU .((((((((((----------------((.((((.(((.....))).)))).)((((((...((.((((..........))))..)).......)))))).))))))))---))).... ( -27.80) >DroWil_CAF1 66836 119 - 1 UUCUCAUAUUAAUAUGCAUGUAUGUAUGUAUAAAACACAUUAAGCGCUUUAAGGCACAAAAUGCUGCCAUAAAAAUCGUCUGCUCGUUAAUUUCUUGUAUACUUGUGUUUUUUGUAUUU ............((((((((....))))))))....(((..((((((((((.((((........)))).))))....((.(((.............))).))..))))))..))).... ( -20.52) >DroYak_CAF1 77392 100 - 1 AACGCACACAA----------------ACAUAAAUCGCAUUAAGCGCUUUAAGGCACAAAAUACUGCCAUAAAAAUCGUUUGCUCGUUAAUUUCUUGUGUAUUUGUGUG---CGUGCUU .((((((((((----------------((((((..(((.....))).((((.((((........)))).)))).....................))))))..)))))))---))).... ( -27.70) >DroAna_CAF1 62780 98 - 1 AA--CACACUC----------------ACAUAAAUCGCAUUAAGCGCUUUAAGGCACAAAAUGCUGCCAUAAAAAUCGUUUGCUCGUUAAUUUCUUGUGUACUUGUGUG---CGUGUUU ..--.((((.(----------------((((((..((((.((((((.((((.((((........)))).))))...)))))).............))))...)))))))---.)))).. ( -22.64) >consensus AACGCACACAA________________ACAUAAAUCGCAUUAAGCGCUUUAAGGCACAAAAUACUGCCAUAAAAAUCGUUUGCUCGUUAAUUUCUUGUGUACUUGUGUG___CGUGCUU ...........................((((((..((((.((((((.((((.((((........)))).))))...)))))).............))))...))))))........... (-10.49 = -10.82 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:02 2006