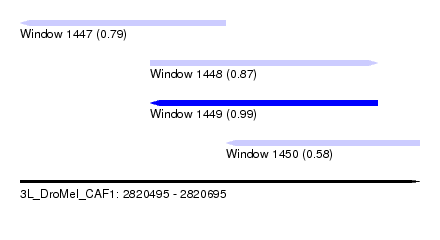
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,820,495 – 2,820,695 |
| Length | 200 |
| Max. P | 0.987366 |

| Location | 2,820,495 – 2,820,598 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.56 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -12.04 |
| Energy contribution | -14.28 |
| Covariance contribution | 2.24 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.792454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2820495 103 - 23771897 GUCA--CCCAAAAAGGGCAACAAUUGUUGCUCUUAAUUCGAACGAUAAGCUUUCGAUCGAAGGAUAUGU-----GUUUAGCCAAA------UUGUAAGAGAUCUGGAAUUUCCUUG---- ....--.((...(((((((((....)))))))))..(((((.(((.......))).)))))))......-----......(((.(------((......))).)))..........---- ( -24.30) >DroSec_CAF1 65449 100 - 1 GUCA--CCCAAAAAGGGCAACAAUUGUUGCUCUUAAUUCGCCAGAUAAGGUUUCGAUCGAAGGAUAUGU-----AAUUACCCAAA-------------AGAUCUGAGUUUUCCUUGGGAA ....--(((((.(((((((((....))))))))).....(((((((..(((..(((((....))).)).-----....)))....-------------..))))).)).....))))).. ( -28.00) >DroSim_CAF1 56949 100 - 1 GUCA--CCCAAAAAGGGCAACAAUUGUUGCUCUUAAUUCGAACGAUAAGGUUUCGAUCGAAGGAUGUGU-----GUUUACCCAAA-------------AGAUCUGAAUUUUCAUUGGGAA ..((--(((...(((((((((....)))))))))..(((((.(((.......))).)))))))..))).-----.....((((((-------------((((....)))))..))))).. ( -28.20) >DroEre_CAF1 68147 107 - 1 GUCA--CCCAAAA-AGGCAGCAAUUGUUACUCUUAAUUUGCCCGAUAAGGCUUCGAUCGAAAGAUAUAUGUAUUGUUUACCCAAAAGAUACAUAUAGAAGAUUUAGAUUU---------- (((.--.......-.((((..(((((.......)))))))))...((((.((((..((....))((((((((((.(((....))).)))))))))))))).)))))))..---------- ( -21.40) >DroYak_CAF1 68036 114 - 1 GUCAUCCCCAAAA-AGGCAACAAUUGUUGCUCUUAAUUUGCCCAAUAAGGUUUCGAUCGAAAGAUGUAU-----GUUUACUUAAAAGAUAAAUGUAAAUGAUCUGGAUUUUCCUGGGGAA ....((((((..(-(((((((....))))).))).......(((...........(((....))).(((-----(((((((....)).)))))))).......))).......)))))). ( -27.50) >consensus GUCA__CCCAAAAAGGGCAACAAUUGUUGCUCUUAAUUCGCCCGAUAAGGUUUCGAUCGAAGGAUAUGU_____GUUUACCCAAA_______U_UA__AGAUCUGGAUUUUCCUUGGGAA ......(((((.(((((((((....)))))))))((((((..(((.......)))(((....)))......................................))))))....))))).. (-12.04 = -14.28 + 2.24)

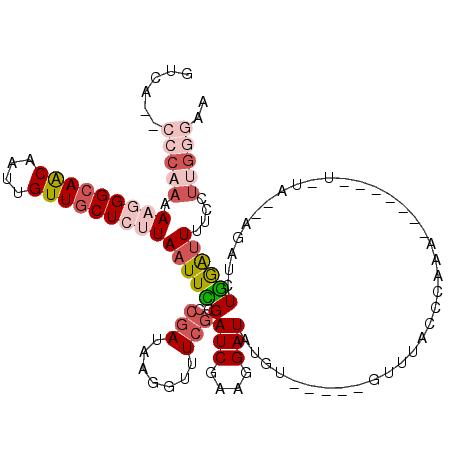
| Location | 2,820,560 – 2,820,674 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -8.40 |
| Energy contribution | -9.60 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.31 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
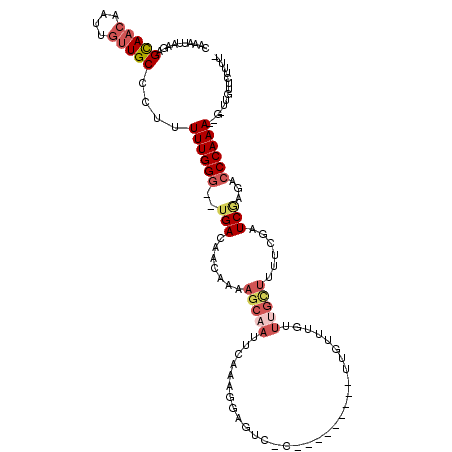
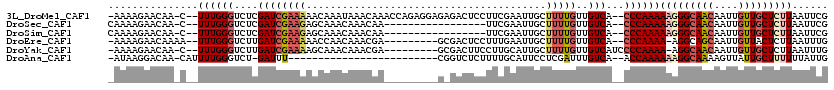
>3L_DroMel_CAF1 2820560 114 + 23771897 CGAAUUAAGAGCAACAAUUGUUGCCCUUUUUGGG--UGACAACAAAAGCAAUUCGAAGGAGUCUCUCCUCUGGUUUGUUUAUUUGUUUUUCGAUCGAGACCCAAA--G-UUGUUCUUUU- ......(((((((((..((((..(((.....)))--..))))...((((((..((.(((((...))))).))..)))))).((((..((((....))))..))))--)-))))))))..- ( -38.70) >DroSec_CAF1 65511 98 + 1 CGAAUUAAGAGCAACAAUUGUUGCCCUUUUUGGG--UGACAACAAAAGCAAUUCGAA-----------------UUGUUUGUUUGCUCUUCGAUCGAGACCCAAA--G-UUGUUCUUUUG ......(((((((((..((((..(((.....)))--..))))..........(((((-----------------..((......))..)))))............--)-))))))))... ( -28.60) >DroSim_CAF1 57011 98 + 1 CGAAUUAAGAGCAACAAUUGUUGCCCUUUUUGGG--UGACAACAAAAGCAAUUCGAA-----------------UUGUUUGUUUGCUCUUCGAUCGAGACCCAAA--G-UUGUUCUUUUG ......(((((((((..((((..(((.....)))--..))))..........(((((-----------------..((......))..)))))............--)-))))))))... ( -28.60) >DroEre_CAF1 68217 105 + 1 CAAAUUAAGAGUAACAAUUGCUGCCU-UUUUGGG--UGACAACAAAAGCAAUUCAAAGGAGUCGC---------UCGUUUGUUGGUUUUUCGAUCAAGACCCAAA--UUUUGUUCUUUU- ......(((((.(((((.........-.((((((--(..((((((((((.((((....)))).))---------)..)))))))......(......))))))))--..))))))))))- ( -22.00) >DroYak_CAF1 68111 106 + 1 CAAAUUAAGAGCAACAAUUGUUGCCU-UUUUGGGGAUGACAACAAAAGCAAUGCAAGGAAGUCGC---------UCGUUUGUUUGCUUUUCGAUCAAGACCCAAA--G-UUGUUCUUUU- ......(((((((((...........-.((((((..(((....((((((((.(((((((......---------)).)))))))))))))...)))...))))))--)-))))))))..- ( -26.80) >DroAna_CAF1 54428 90 + 1 CAAUAAAAAAGCAAUAACUUUUGCCUUUUUUGGU--UGACAAAUCGAGGAAUGCAAAAGAGACCG-------------------------AAAUC-AGACCCAAAAUG-UUGUCCUUAU- ..........((((((.(((((((.(((((((((--(....)))))))))).))))))).((...-------------------------...))-..........))-))))......- ( -19.20) >consensus CAAAUUAAGAGCAACAAUUGUUGCCCUUUUUGGG__UGACAACAAAAGCAAUUCAAAGGAGUC_C_________UUGUUUGUUUGCUUUUCGAUCGAGACCCAAA__G_UUGUUCUUUU_ ..........(((((....)))))....((((((..(((.......(((((...............................)))))......)))...))))))............... ( -8.40 = -9.60 + 1.20)
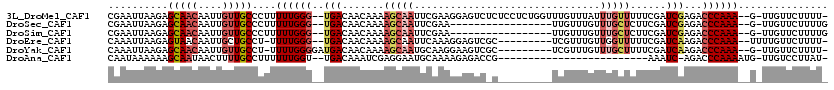
| Location | 2,820,560 – 2,820,674 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.22 |
| Mean single sequence MFE | -25.63 |
| Consensus MFE | -11.20 |
| Energy contribution | -12.28 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987366 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
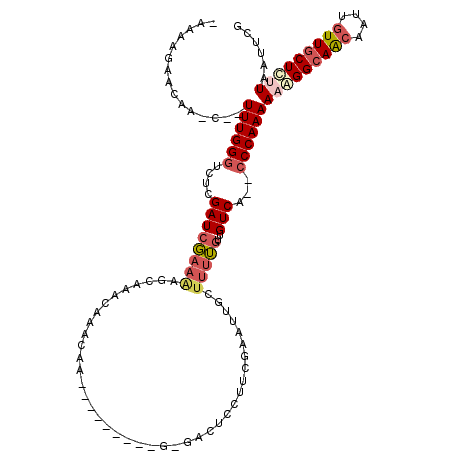
>3L_DroMel_CAF1 2820560 114 - 23771897 -AAAAGAACAA-C--UUUGGGUCUCGAUCGAAAAACAAAUAAACAAACCAGAGGAGAGACUCCUUCGAAUUGCUUUUGUUGUCA--CCCAAAAAGGGCAACAAUUGUUGCUCUUAAUUCG -....(((...-.--(((((((...(((.....((((((....(((..(..(((((...)))))..)..)))..))))))))))--))))))(((((((((....)))))))))..))). ( -30.80) >DroSec_CAF1 65511 98 - 1 CAAAAGAACAA-C--UUUGGGUCUCGAUCGAAGAGCAAACAAACAA-----------------UUCGAAUUGCUUUUGUUGUCA--CCCAAAAAGGGCAACAAUUGUUGCUCUUAAUUCG .....(((...-.--(((((((..((((..((((((((........-----------------......))))))))))))..)--))))))(((((((((....)))))))))..))). ( -29.84) >DroSim_CAF1 57011 98 - 1 CAAAAGAACAA-C--UUUGGGUCUCGAUCGAAGAGCAAACAAACAA-----------------UUCGAAUUGCUUUUGUUGUCA--CCCAAAAAGGGCAACAAUUGUUGCUCUUAAUUCG .....(((...-.--(((((((..((((..((((((((........-----------------......))))))))))))..)--))))))(((((((((....)))))))))..))). ( -29.84) >DroEre_CAF1 68217 105 - 1 -AAAAGAACAAAA--UUUGGGUCUUGAUCGAAAAACCAACAAACGA---------GCGACUCCUUUGAAUUGCUUUUGUUGUCA--CCCAAAA-AGGCAGCAAUUGUUACUCUUAAUUUG -..((((......--....(((.((......)).)))(((((..((---------((((.((....)).))))))((((((((.--.......-.)))))))))))))..))))...... ( -19.60) >DroYak_CAF1 68111 106 - 1 -AAAAGAACAA-C--UUUGGGUCUUGAUCGAAAAGCAAACAAACGA---------GCGACUUCCUUGCAUUGCUUUUGUUGUCAUCCCCAAAA-AGGCAACAAUUGUUGCUCUUAAUUUG -..........-.--((((((...((((..((((((((.....(((---------(.(....)))))..))))))))...))))..))))))(-(((((((....))))).)))...... ( -26.20) >DroAna_CAF1 54428 90 - 1 -AUAAGGACAA-CAUUUUGGGUCU-GAUUU-------------------------CGGUCUCUUUUGCAUUCCUCGAUUUGUCA--ACCAAAAAAGGCAAAAGUUAUUGCUUUUUUAUUG -...((((...-((....(((.((-(....-------------------------))).)))...))...))))..........--....((((((((((......)))))))))).... ( -17.50) >consensus _AAAAGAACAA_C__UUUGGGUCUCGAUCGAAAAGCAAACAAACAA_________G_GACUCCUUCGAAUUGCUUUUGUUGUCA__CCCAAAAAAGGCAACAAUUGUUGCUCUUAAUUCG ...............((((((....((((((((........................................)))))..)))...))))))(((((((((....)))))))))...... (-11.20 = -12.28 + 1.08)
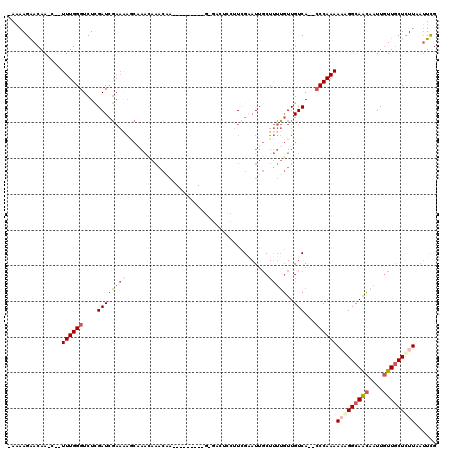
| Location | 2,820,598 – 2,820,695 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 83.10 |
| Mean single sequence MFE | -19.77 |
| Consensus MFE | -12.90 |
| Energy contribution | -13.01 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2820598 97 - 23771897 AAUCGAAAACAAAGGCAGGCCAAAAGAACAA-CUUUGGGUCUCGAUCGAAAAACAAAUAAACAAACCAGAGGAGAGACUCCUUCGAAUUGCUUUUGUU .......((((((((((((((.((((.....-)))).)))).........................(..(((((...)))))..)...)))))))))) ( -23.90) >DroEre_CAF1 68254 89 - 1 AAUCGAAAACAAAGGCCAGCCAAAAGAACAAAAUUUGGGUCUUGAUCGAAAAACCAACAAACGA---------GCGACUCCUUUGAAUUGCUUUUGUU ..((((.....((((((.....(((........))).))))))..))))......(((((..((---------((((.((....)).))))))))))) ( -16.40) >DroYak_CAF1 68150 88 - 1 AUUCGAAAACAAAGGCAAGCCAAAAGAACAA-CUUUGGGUCUUGAUCGAAAAGCAAACAAACGA---------GCGACUUCCUUGCAUUGCUUUUGUU .......(((((((((((((..((((.....-))))(((..(((.(((.............)))---------.)))..)))..)).))))))))))) ( -19.02) >consensus AAUCGAAAACAAAGGCAAGCCAAAAGAACAA_CUUUGGGUCUUGAUCGAAAAACAAACAAACGA_________GCGACUCCUUUGAAUUGCUUUUGUU .......(((((((((((.(((((.........)))))(((..................................))).........))))))))))) (-12.90 = -13.01 + 0.11)

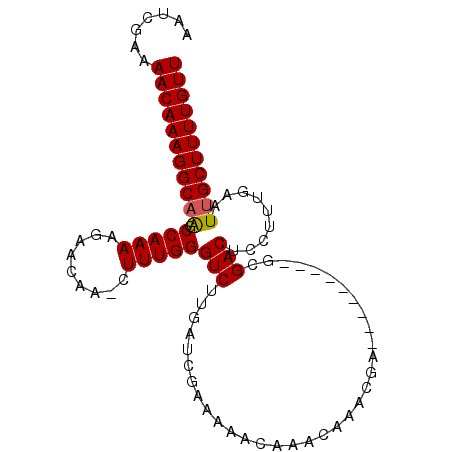
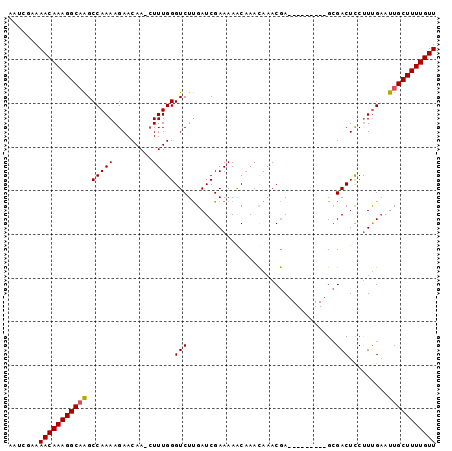
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:55:00 2006