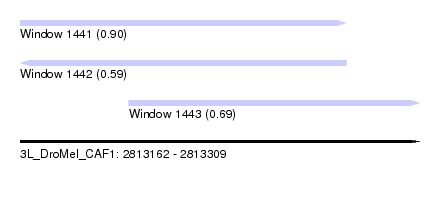
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,813,162 – 2,813,309 |
| Length | 147 |
| Max. P | 0.898587 |

| Location | 2,813,162 – 2,813,282 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -22.54 |
| Consensus MFE | -22.24 |
| Energy contribution | -21.92 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2813162 120 + 23771897 UAUUCCAAUCAUUCUUUGAACACAAUAUGGAUUGGAUUGGCCUUAACUAAUUCCAAAAUGAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACAC ...(((((((.....(((....)))....)))))))..((((.................).)))(((.((.((((.((((.....((((.....))))))))..)))).))..))).... ( -22.83) >DroSec_CAF1 58210 120 + 1 UAUUCCAAUCAUUCUUUGAACACAAUAUGGAUUGGAUUGGCCUUAACUAAUUCCAAAAUGAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACAC ...(((((((.....(((....)))....)))))))..((((.................).)))(((.((.((((.((((.....((((.....))))))))..)))).))..))).... ( -22.83) >DroSim_CAF1 49502 120 + 1 UAUUCCAAUCAUUCUUUGAACACAAUAUGGAUUGGAUUGGCCUUAACUAAUUCCAAAAUGAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACAC ...(((((((.....(((....)))....)))))))..((((.................).)))(((.((.((((.((((.....((((.....))))))))..)))).))..))).... ( -22.83) >DroEre_CAF1 60818 120 + 1 UAUUUCAAUCAUUCUUUGAACACAAUAUGGAUUGGAUUGGCCUUAACUAAUUCCGAAAUGAGCCGAAUCAAAUCCCUUGGCCUCGACGUACAUCACGUCCAAAUGGAUAUGUUUUCACAC ...(((((((.....(((....)))....))))))).((.....(((....((((....(((((((..........)))).)))(((((.....)))))....))))...)))....)). ( -21.80) >DroYak_CAF1 60218 120 + 1 UAUUCCAAUCAUUCUUUGAGCACAAUAUGGAUUGGAUUGGCCUUAACUAAUUCCAAAAUAAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACAC ...(((((((.....(((....)))....)))))))..(((.(((.............))))))(((.((.((((.((((.....((((.....))))))))..)))).))..))).... ( -22.42) >consensus UAUUCCAAUCAUUCUUUGAACACAAUAUGGAUUGGAUUGGCCUUAACUAAUUCCAAAAUGAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACAC ...(((((((.....(((....)))....)))))))..(((.(((.............))))))(((.((.((((.((((.....((((.....))))))))..)))).))..))).... (-22.24 = -21.92 + -0.32)

| Location | 2,813,162 – 2,813,282 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -27.88 |
| Energy contribution | -27.76 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2813162 120 - 23771897 GUGUGAAAACAUAUCCAUUUGGACGUGAUGUACGUUGAGGCCAAGGGAUUUGAUUCGGCUCAUUUUGGAAUUAGUUAAGGCCAAUCCAAUCCAUAUUGUGUUCAAAGAAUGAUUGGAAUA ((((....)))).((((((((((((..((((...(((....))).(((((.((((.((((.(((........)))...)))))))).)))))))))..)))))))).......))))... ( -30.51) >DroSec_CAF1 58210 120 - 1 GUGUGAAAACAUAUCCAUUUGGACGUGAUGUACGUUGAGGCCAAGGGAUUUGAUUCGGCUCAUUUUGGAAUUAGUUAAGGCCAAUCCAAUCCAUAUUGUGUUCAAAGAAUGAUUGGAAUA ((((....)))).((((((((((((..((((...(((....))).(((((.((((.((((.(((........)))...)))))))).)))))))))..)))))))).......))))... ( -30.51) >DroSim_CAF1 49502 120 - 1 GUGUGAAAACAUAUCCAUUUGGACGUGAUGUACGUUGAGGCCAAGGGAUUUGAUUCGGCUCAUUUUGGAAUUAGUUAAGGCCAAUCCAAUCCAUAUUGUGUUCAAAGAAUGAUUGGAAUA ((((....)))).((((((((((((..((((...(((....))).(((((.((((.((((.(((........)))...)))))))).)))))))))..)))))))).......))))... ( -30.51) >DroEre_CAF1 60818 120 - 1 GUGUGAAAACAUAUCCAUUUGGACGUGAUGUACGUCGAGGCCAAGGGAUUUGAUUCGGCUCAUUUCGGAAUUAGUUAAGGCCAAUCCAAUCCAUAUUGUGUUCAAAGAAUGAUUGAAAUA ((((....))))...(((((.((((((...))))))(((..(((.(((((.((((.((((.....(.......)....)))))))).)))))...)))..)))...)))))......... ( -29.10) >DroYak_CAF1 60218 120 - 1 GUGUGAAAACAUAUCCAUUUGGACGUGAUGUACGUUGAGGCCAAGGGAUUUGAUUCGGCUUAUUUUGGAAUUAGUUAAGGCCAAUCCAAUCCAUAUUGUGCUCAAAGAAUGAUUGGAAUA ((((....)))).((((.((.((((((...))))))(((..(((.(((((.((((.(((((................))))))))).)))))...)))..))).......)).))))... ( -30.69) >consensus GUGUGAAAACAUAUCCAUUUGGACGUGAUGUACGUUGAGGCCAAGGGAUUUGAUUCGGCUCAUUUUGGAAUUAGUUAAGGCCAAUCCAAUCCAUAUUGUGUUCAAAGAAUGAUUGGAAUA ((((....)))).((((.((.((((((...))))))(((..(((.(((((.((((.((((.(((........)))...)))))))).)))))...)))..))).......)).))))... (-27.88 = -27.76 + -0.12)

| Location | 2,813,202 – 2,813,309 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -25.98 |
| Consensus MFE | -22.48 |
| Energy contribution | -22.24 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
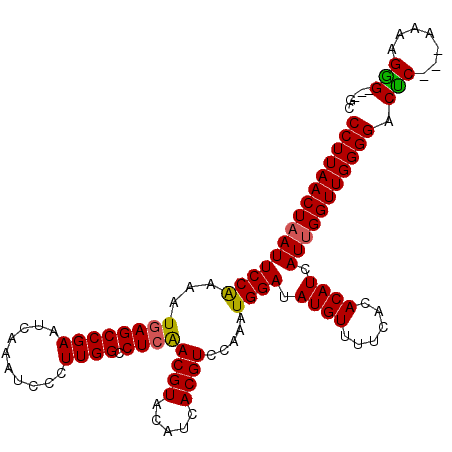
>3L_DroMel_CAF1 2813202 107 + 23771897 CCUUAACUAAUUCCAAAAUGAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACACAUCAUUGGUUGGGGACUC---AAAAGGG---GC (((((((((((((((...((((((((..........)))).))))((((.....)))).....)))).((((......)))).))))))))))).(((---....)))---.. ( -25.00) >DroSec_CAF1 58250 107 + 1 CCUUAACUAAUUCCAAAAUGAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACACAUCAUUGGUUGGGGACUC---AAAAGGG---GC (((((((((((((((...((((((((..........)))).))))((((.....)))).....)))).((((......)))).))))))))))).(((---....)))---.. ( -25.00) >DroSim_CAF1 49542 107 + 1 CCUUAACUAAUUCCAAAAUGAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACACAUCAUUGGUUGGGGACUC---AAAAGGG---GC (((((((((((((((...((((((((..........)))).))))((((.....)))).....)))).((((......)))).))))))))))).(((---....)))---.. ( -25.00) >DroEre_CAF1 60858 110 + 1 CCUUAACUAAUUCCGAAAUGAGCCGAAUCAAAUCCCUUGGCCUCGACGUACAUCACGUCCAAAUGGAUAUGUUUUCACACAUCAUUGGUUGGGGACUC---AAAGGAGGAAGC (((((((((((((((....(((((((..........)))).)))(((((.....)))))....)))).((((......)))).))))))))))).(((---....)))..... ( -27.70) >DroYak_CAF1 60258 113 + 1 CCUUAACUAAUUCCAAAAUAAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACACAUCAUAGGUUGGGGACCCCAAAAGGGGGGAUGC ...........(((....................(((..((((..((((.....)))).......(((.(((....))).)))..))))..))).((((....)))))))... ( -27.20) >consensus CCUUAACUAAUUCCAAAAUGAGCCGAAUCAAAUCCCUUGGCCUCAACGUACAUCACGUCCAAAUGGAUAUGUUUUCACACAUCAUUGGUUGGGGACUC___AAAAGGG___GC (((((((((((((((...((((((((..........)))).))))((((.....)))).....)))).((((......)))).))))))))))).(((.......)))..... (-22.48 = -22.24 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:53 2006