| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,804,844 – 2,804,935 |
| Length | 91 |
| Max. P | 0.660377 |

| Location | 2,804,844 – 2,804,935 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.43 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -13.51 |
| Energy contribution | -13.70 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
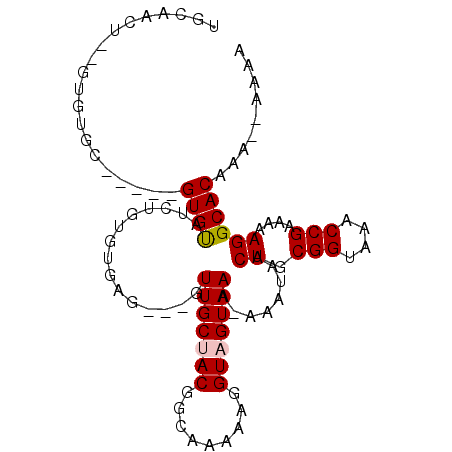
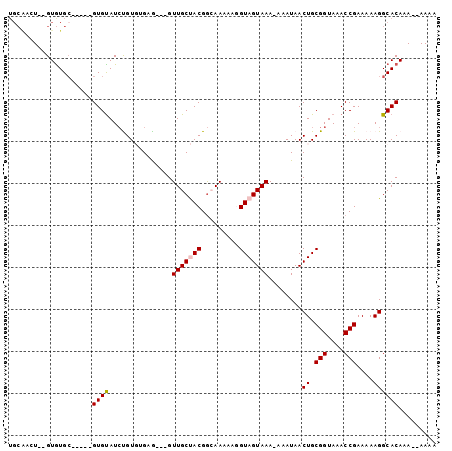
>3L_DroMel_CAF1 2804844 91 + 23771897 UGCGACUUGGUGUGC-----GUGUAUCUGUGUGCG---GUUGCUACGGCAAAAAGGUAGUAAA-AAAUAACUGCGGUAAACCGAAAAAGGCACAAA--AAAA ......((..(((((-----.(...((.((.(((.---.((((....))))....(((((...-.....))))).))).)).))...).)))))..--)).. ( -22.40) >DroPse_CAF1 59185 95 + 1 UGCACGUGUGUGUGCCCGUAGUGUAUCCGU--GAG---GUUGCCACGGCAAAAACGUAGUAAAAAAAUAACUGCGGUAAACCGAAAAAGGCACAAA--AAUA .....((...((((((....(.(((.((..--..)---).)))).(((......((((((.........)))))).....))).....))))))..--.)). ( -26.60) >DroSim_CAF1 39854 93 + 1 UGCAACUUGGUGUGC-----GUGUAUCUGUGUGCG---GUUGCUACGGCAAAAAGGUAGUAAA-AAAUAACUGCGGUAAACCGAAAAAGGCACAAAAAAAAA ......((..(((((-----.(...((.((.(((.---.((((....))))....(((((...-.....))))).))).)).))...).)))))..)).... ( -22.40) >DroMoj_CAF1 51786 82 + 1 -----CU--GUGUGC-----GUGCAUGUGUGAG-A---GUUGCUACGA-AAAAAGGUAGUAAA-AAAUAACUGCGGUAAACCGAAAAAGGCACAAA--AAAU -----..--.(((((-----..(((.((.....-.---.(((((((..-......))))))).-.....)))))((....)).......)))))..--.... ( -20.22) >DroAna_CAF1 39072 91 + 1 UGCAAGU--GUAUGG-----GUGUAUCUGUGUGUGCAUGUUGCUACGGAAAAAAGGUAGUAAA-AAAUAACUGCGGUAAACCGAAAAAGGCACAAA--AAA- .....((--((.(.(-----((....(((((.((.....(((((((.........))))))).-.....)))))))...))).).....))))...--...- ( -18.40) >DroPer_CAF1 59610 95 + 1 UGCACGUGUGUGUGCCCGUAGUGUAUCCGU--GAG---GUUGCCACGGCAAAAACGUAGUAAAAAAAUAACUGCGGUAAACCGAAAAAGGCACAAA--AAUA .....((...((((((....(.(((.((..--..)---).)))).(((......((((((.........)))))).....))).....))))))..--.)). ( -26.60) >consensus UGCAACU__GUGUGC_____GUGUAUCUGUGUGAG___GUUGCUACGGCAAAAAGGUAGUAAA_AAAUAACUGCGGUAAACCGAAAAAGGCACAAA__AAAA ....................((((...............(((((((.........)))))))........((.(((....)))....))))))......... (-13.51 = -13.70 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:47 2006