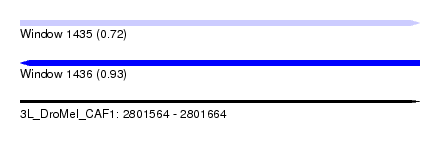
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,801,564 – 2,801,664 |
| Length | 100 |
| Max. P | 0.932291 |

| Location | 2,801,564 – 2,801,664 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -22.19 |
| Consensus MFE | -18.57 |
| Energy contribution | -18.23 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.724644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2801564 100 + 23771897 UU---UACUGUUUUUUAUGUCGUGCCAUUUCAGCGAAGCUCGGCAACAAAGCAUAAUAAAACUAAUUGUCUGCGCUUCGCUCUCACACACAGAUACACCCAUA ..---............(((((((.......((((((((..(....)...(((..((((......)))).)))))))))))......))).))))........ ( -20.52) >DroSec_CAF1 46657 100 + 1 UU---UACUGUUUUUUAUGUCGUGCCAUUUCAGCGAAGCUCGGCAACAAAGCAUAAUAAAACUAAUUGUCUGCGCUUCGCUCUCACACACAGAUACACCCAUA ..---............(((((((.......((((((((..(....)...(((..((((......)))).)))))))))))......))).))))........ ( -20.52) >DroSim_CAF1 36581 100 + 1 UU---UACUGUUUUUUAUGUCGUGCCAUUUCAGCGAAGCUCGGCAACAAAGCAUAAUAAAACUAAUUGUCUGCGCUUCGCUCUCACACACAGAUGCACCCAUA ..---..........((((..((((.((((.((((((((..(....)...(((..((((......)))).))))))))))).........)))))))).)))) ( -20.90) >DroEre_CAF1 48820 101 + 1 UUU--UACUGUUUUUUAUGUCGUGCCAUUUCAGCGAAGCUCGGCAACAAAGCAUAAUAAAACUAAUUGUCUGCGCUUCGCUCUCACACACAGAUACACCCAUA ...--............(((((((.......((((((((..(....)...(((..((((......)))).)))))))))))......))).))))........ ( -20.52) >DroYak_CAF1 48183 103 + 1 UUUUGUACUGUUUUUUAUGUCGUGCCAUUUCAGCGAAGCUCGGCAACAAAGCAUAAUAAAACUAAUUGUCUGCGCUUCGCUCUCACACACAGAUACACCCAUA ...((((((((.......((.(((.......((((((((..(....)...(((..((((......)))).)))))))))))..))))))))).))))...... ( -21.21) >DroMoj_CAF1 47253 101 + 1 GUUGUUGUUGCGUUUUAUGUCGUGCCAUUUCAGCGAAGCUC-GCAACAAAGCAUAAUAAAACUAAUUGUCUGCGCUUCGCUCUCGCACACACA-GCGCACAAA ....((((.(((((...(((.((((......((((((((..-(((((((................)))).)))))))))))...))))))).)-)))))))). ( -29.49) >consensus UU___UACUGUUUUUUAUGUCGUGCCAUUUCAGCGAAGCUCGGCAACAAAGCAUAAUAAAACUAAUUGUCUGCGCUUCGCUCUCACACACAGAUACACCCAUA ........(((......(((.(((.......((((((((..(....)...(((..((((......)))).)))))))))))....))))))...)))...... (-18.57 = -18.23 + -0.33)

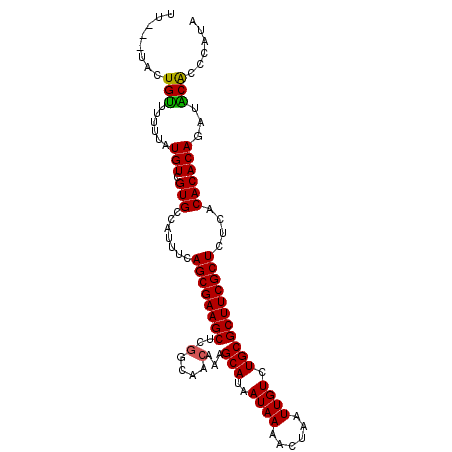
| Location | 2,801,564 – 2,801,664 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 93.79 |
| Mean single sequence MFE | -26.78 |
| Consensus MFE | -24.90 |
| Energy contribution | -24.48 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2801564 100 - 23771897 UAUGGGUGUAUCUGUGUGUGAGAGCGAAGCGCAGACAAUUAGUUUUAUUAUGCUUUGUUGCCGAGCUUCGCUGAAAUGGCACGACAUAAAAAACAGUA---AA ..((..(.....(((((((.(.((((((((...(.((((.(((........)))..)))).)..))))))))....).)))).)))....)..))...---.. ( -24.50) >DroSec_CAF1 46657 100 - 1 UAUGGGUGUAUCUGUGUGUGAGAGCGAAGCGCAGACAAUUAGUUUUAUUAUGCUUUGUUGCCGAGCUUCGCUGAAAUGGCACGACAUAAAAAACAGUA---AA ..((..(.....(((((((.(.((((((((...(.((((.(((........)))..)))).)..))))))))....).)))).)))....)..))...---.. ( -24.50) >DroSim_CAF1 36581 100 - 1 UAUGGGUGCAUCUGUGUGUGAGAGCGAAGCGCAGACAAUUAGUUUUAUUAUGCUUUGUUGCCGAGCUUCGCUGAAAUGGCACGACAUAAAAAACAGUA---AA ((((.((((.((.......)).((((((((...(.((((.(((........)))..)))).)..))))))))......))))..))))..........---.. ( -26.20) >DroEre_CAF1 48820 101 - 1 UAUGGGUGUAUCUGUGUGUGAGAGCGAAGCGCAGACAAUUAGUUUUAUUAUGCUUUGUUGCCGAGCUUCGCUGAAAUGGCACGACAUAAAAAACAGUA--AAA ..((..(.....(((((((.(.((((((((...(.((((.(((........)))..)))).)..))))))))....).)))).)))....)..))...--... ( -24.50) >DroYak_CAF1 48183 103 - 1 UAUGGGUGUAUCUGUGUGUGAGAGCGAAGCGCAGACAAUUAGUUUUAUUAUGCUUUGUUGCCGAGCUUCGCUGAAAUGGCACGACAUAAAAAACAGUACAAAA ......((((.((((((((.(.((((((((...(.((((.(((........)))..)))).)..))))))))....).))))..........))))))))... ( -26.80) >DroMoj_CAF1 47253 101 - 1 UUUGUGCGC-UGUGUGUGCGAGAGCGAAGCGCAGACAAUUAGUUUUAUUAUGCUUUGUUGC-GAGCUUCGCUGAAAUGGCACGACAUAAAACGCAACAACAAC .(((((((.-(((((((((.(.((((((((...(((((..(((........))))))))..-..))))))))....).)))).)))))...))).)))).... ( -34.20) >consensus UAUGGGUGUAUCUGUGUGUGAGAGCGAAGCGCAGACAAUUAGUUUUAUUAUGCUUUGUUGCCGAGCUUCGCUGAAAUGGCACGACAUAAAAAACAGUA___AA .....((.....(((((((.(.((((((((...(((((..(((........)))))))).....))))))))....).)))).)))......))......... (-24.90 = -24.48 + -0.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:46 2006