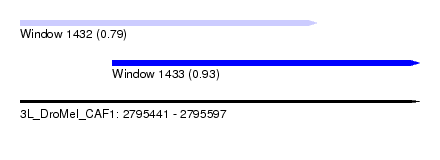
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,795,441 – 2,795,597 |
| Length | 156 |
| Max. P | 0.932758 |

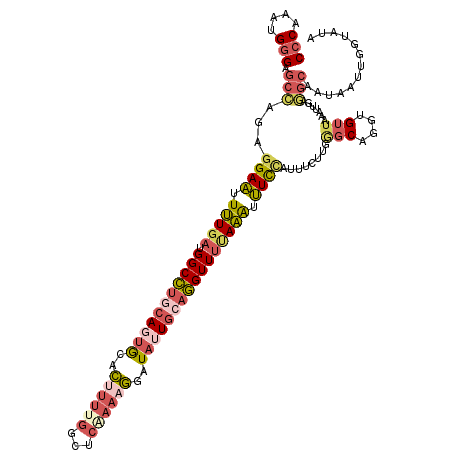
| Location | 2,795,441 – 2,795,557 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -31.06 |
| Consensus MFE | -23.26 |
| Energy contribution | -24.43 |
| Covariance contribution | 1.18 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2795441 116 + 23771897 UGUUUGCAAUCGAUAAGCCAGGCUAA-AACUUAUGCACCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAGUGCACUUUUGGCUCUAAAGAAUAUUGCAGGUUUUAAAUUUCCA .((((((....).)))))..((((..-..(....)..(((....)))))))...((((.(((((.(((((((((((..((((.......))))..)))))))))))))))).)))). ( -32.90) >DroSec_CAF1 40661 117 + 1 UGUUUGCAAUCGAUAAGCCAGGCUAAAAACUUAUGCACCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAGUGCACUUUUGGCUCGAAAGGAUAUUGCAGGUUUUAAAUUUCCA .((((((....).)))))..((((.....(....)..(((....)))))))...((((.(((((.(((((((((((..((((((...))))))..)))))))))))))))).)))). ( -37.00) >DroSim_CAF1 30513 117 + 1 UGUUUGCAAUCGAUAAGCCAGGCUAAAAACUUAUGCACCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAGUGCACUUUUGGCUCGAAAGGAUAUUGCAGGUUUUAAAUUUCCA .((((((....).)))))..((((.....(....)..(((....)))))))...((((.(((((.(((((((((((..((((((...))))))..)))))))))))))))).)))). ( -37.00) >DroEre_CAF1 42573 116 + 1 UGUUUGCAGACGAUAAGCUGGGCCAA-AACUUAUGCACCCAAAUGGGAACCAGAGGAAUUUUGAUGGCCUGCAUUGCACAUUUGGCUCAAAAGGAUACUGCAGGUUUUAAAUUUCAA ..(((((((.........((((((((-(....((((((((....)))..(((..((....))..)))..)))))......)))))))))........)))))))............. ( -28.53) >DroYak_CAF1 41935 116 + 1 UGUUUGCAAACGAUAAGCUAGGCUAA-AACUUAUGCACCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAUUGCACUUUUGGCUCAAAAGGAUAUUGAAGGUUUCAAACUUCCA .....((.........((((((....-..)))).)).(((....))).))....((((.(((((.(((((.((.((..((((((...))))))..)).)).)))))))))).)))). ( -29.60) >DroAna_CAF1 30502 99 + 1 UGUU-GGAAUCGAUUAGUCGG------AGCUCUGGCUCCCA--UGAGAGUUCAAGGACCUAU--UGGCAGACAGGACAUUUUUUC-------AGUUAUUUCAUGUUUGAUGUGUUUA ((((-....(((((..(((.(------((((((.(......--).)))))))...)))..))--)))..))))((((((....((-------((...........)))).)))))). ( -21.30) >consensus UGUUUGCAAUCGAUAAGCCAGGCUAA_AACUUAUGCACCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAGUGCACUUUUGGCUCAAAAGGAUAUUGCAGGUUUUAAAUUUCCA ..................................((.(((....))).))....((((.(((((.(((((((((((..((((((...))))))..)))))))))))))))).)))). (-23.26 = -24.43 + 1.18)

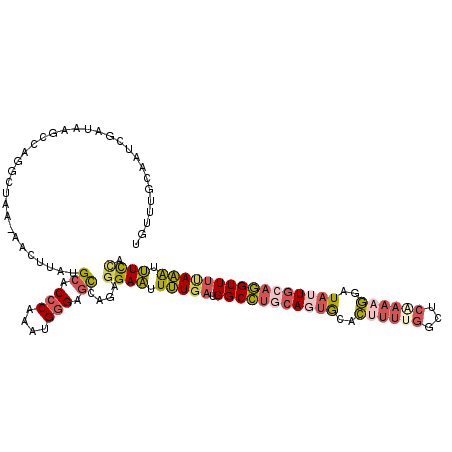
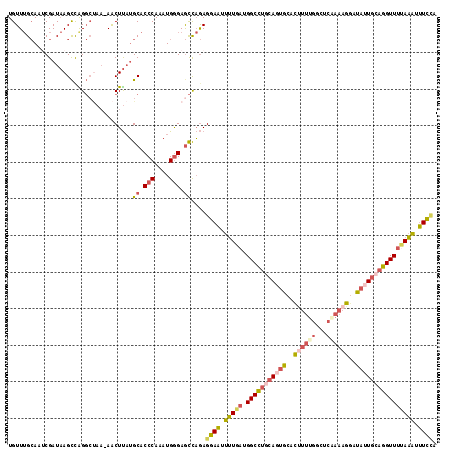
| Location | 2,795,477 – 2,795,597 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.17 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -21.83 |
| Energy contribution | -23.57 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2795477 120 + 23771897 CCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAGUGCACUUUUGGCUCUAAAGAAUAUUGCAGGUUUUAAAUUUCCAUUUCUUGGGCAGGUGUUAAAUUGAGGCAAUAAUUGGCGUA (((....))).(((((.((((.(((((.(((((((((((..((((.......))))..)))))))))))))))).)))).........((.(((.....)))...)).....)))))... ( -36.90) >DroSec_CAF1 40698 120 + 1 CCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAGUGCACUUUUGGCUCGAAAGGAUAUUGCAGGUUUUAAAUUUCCAUUUCUUGGGCAGGUGUUAAAUUGAUGCAAUAAUUGUUAUA (((....))).((((((((((.(((((.(((((((((((..((((((...))))))..)))))))))))))))).))))...)))..)))..(((((.....)))))............. ( -37.80) >DroSim_CAF1 30550 120 + 1 CCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAGUGCACUUUUGGCUCGAAAGGAUAUUGCAGGUUUUAAAUUUCCAUUUCUUGAGCAGGUGUUAAAUUGAGGCAAUAAUUGUUAUA (((....))).(((...((((.(((((.(((((((((((..((((((...))))))..)))))))))))))))).))))...((...(((....))).....)))))............. ( -38.00) >DroEre_CAF1 42609 120 + 1 CCCAAAUGGGAACCAGAGGAAUUUUGAUGGCCUGCAUUGCACAUUUGGCUCAAAAGGAUACUGCAGGUUUUAAAUUUCAAUUUCUUGGCCAGGUGUUAAAUUGAGGCAAUCAUUGGCAUA (((....))).......(((((((....((((((((.((..(.((((...)))).)..)).))))))))..))))))).........(((((.((((.......))))....)))))... ( -30.20) >DroYak_CAF1 41971 119 + 1 CCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAUUGCACUUUUGGCUCAAAAGGAUAUUGAAGGUUUCAAACUUCCAAUU-UUGGGCAGGUGUUAAAUUGAGGCAAUAAUUGGUAUG (((....))).(((((.((((.(((((.(((((.((.((..((((((...))))))..)).)).)))))))))).))))....-....((.(((.....)))...)).....)))))... ( -30.90) >DroAna_CAF1 30532 90 + 1 CCCA--UGAGAGUUCAAGGACCUAU--UGGCAGACAGGACAUUUUUUC-------AGUUAUUUCAUGUUUGAUGUGUUUAUUGACU----------UUUAUUUAACCAAAU--------- ...(--(((((((.(((((((.(((--..(((((...(((........-------.)))...)).)).)..))).)))).))))))----------)))))..........--------- ( -13.60) >consensus CCCAAAUGGGAGCCAGAGGAAUUUUGAUGGCCUGCAGUGCACUUUUGGCUCAAAAGGAUAUUGCAGGUUUUAAAUUUCCAUUUCUUGGGCAGGUGUUAAAUUGAGGCAAUAAUUGGUAUA (((....))).(((...((((.(((((.(((((((((((..((((((...))))))..)))))))))))))))).))))........(((....))).......)))............. (-21.83 = -23.57 + 1.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:44 2006