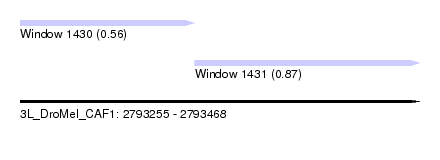
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,793,255 – 2,793,468 |
| Length | 213 |
| Max. P | 0.871833 |

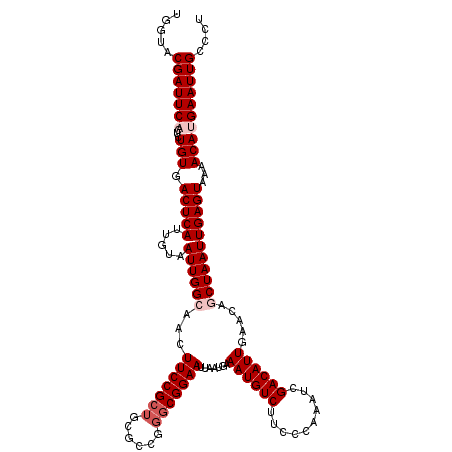
| Location | 2,793,255 – 2,793,348 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.02 |
| Mean single sequence MFE | -22.12 |
| Consensus MFE | -14.23 |
| Energy contribution | -15.10 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2793255 93 + 23771897 -----AC---------AUACAUAAAAGUAAAUCUAAUUAAACCAGAAUGCCUCACGAUCCGAGGUCUUCGAAUAGAACCUGU------GUUCUAUAUCUACAUGAGGGGGAAC -----..---------.(((......)))............((.(((.(((((.......))))).)))(((((((((....------))))))).)).......))...... ( -18.20) >DroSec_CAF1 38497 96 + 1 -----AC---------AUACAUAAAAGUAAAUCUAAUUAAACCAGAAUGCCUCACGAUCCGAGGUCUUCGAACAGAACCUGUGUAAAUGUUCUAUAUUUACG---AGGGGAAC -----..---------.(((......)))............((.(((.(((((.......))))).)))........(((.(((((((((...)))))))))---)))))... ( -20.30) >DroSim_CAF1 28331 96 + 1 -----AC---------AUACAUAAAAGUAAAUCUAAUUAAACCAGAAUGCCUCACGAUCCGAGGUCUUCGAAUAGAACCUGUGUAAAUGUUCUAUAUUUACG---AGGGGAAC -----..---------.(((......)))............((.(((.(((((.......))))).)))........(((.(((((((((...)))))))))---)))))... ( -20.30) >DroEre_CAF1 40385 110 + 1 UUUAGCCCUACAUACAAUACAUAAAAGUAAAUCUAAUUAAACGAGAAUGCCCUGCGAUCCGAGGCCUUCGAAUAGAACCUGUGUAAAUGGCCUAUGGUUGCA---GGGGGAAU ......((.........(((......)))....................((((((((((..(((((.....((((...))))......)))))..)))))))---)))))... ( -29.70) >consensus _____AC_________AUACAUAAAAGUAAAUCUAAUUAAACCAGAAUGCCUCACGAUCCGAGGUCUUCGAAUAGAACCUGUGUAAAUGUUCUAUAUUUACA___AGGGGAAC .................(((......)))..(((..........(((.(((((.......))))).)))........(((.((((((((.....))))))))...)))))).. (-14.23 = -15.10 + 0.87)

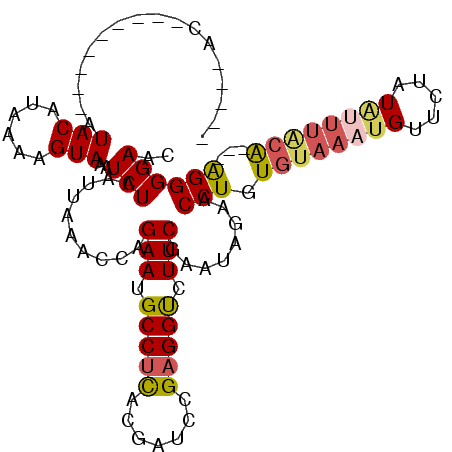
| Location | 2,793,348 – 2,793,468 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.33 |
| Mean single sequence MFE | -30.70 |
| Consensus MFE | -26.22 |
| Energy contribution | -27.42 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2793348 120 + 23771897 UGGUACGAUUCACUUUGUGACUCAUUGUAAUUGGCAACUUCCGCUGCGCCGGGCGGAAUUAAUGAAAUGUCUUCCCAAAUCGACAUUGAACAGCUAAUUGAGUAAAACAUGAAUUGCCCU .((..(((((((...(((.(((((.....((((((...(((((((......))))))).......((((((..........)))))).....)))))))))))...)))))))))).)). ( -32.70) >DroSec_CAF1 38593 120 + 1 UGGUACGAUUCCCUCUGUGACUCAUUUUAAUUGGCAACUUCCGCUGCGCCGGGCGGAAUUAAUGAAAUGUCUUCUCAAAUCGACAUUGAACAGCUAAUUGAGUAAAACAUGAAUUGCCCC .((..((((((....(((.(((((.....((((((...(((((((......))))))).......((((((..........)))))).....)))))))))))...))).))))))..)) ( -32.10) >DroSim_CAF1 28427 120 + 1 UGGUACGAUUCCCUCUGUGACUCAUUGUAAUUGGCAACUUCCGCUGCGCCGCUCGGAAUUAAUGAAAUGUCUUCUCAAAUCGACAUUGAACAGCUAAUUGAGUAAAACAUGAAUUGCCCU .((..((((((....(((.(((((.....((((((...(((((..((...)).))))).......((((((..........)))))).....)))))))))))...))).)))))).)). ( -26.60) >DroEre_CAF1 40495 120 + 1 UCGUACGAUUCACUCUGUGACUCAUUGUAAUUGGCAACUUCCGCUGCGGCAGGCGGAAUUAAUGAAAUGUCUUCCCAAAUCGACAUUGAACAACUAAUUGAGUAAAACAUGAAUUGCCCC ..(..(((((((...(((.(((((...((.(((.....(((((((......))))))).......((((((..........))))))...))).))..)))))...))))))))))..). ( -29.40) >DroYak_CAF1 39816 120 + 1 UCGUGCGAUUCAGUCUGUGACUCAUUGUAAUUGGCAACUUCCGCUGUGCCAGGCGGAAUUAAUGAAAUGUCUUCCCAAAUCGACAUUGAACAACUAAUUGAGUAAAACAUGAAUUGCCCU ..(.((((((((...(((.(((((...((.(((.....(((((((......))))))).......((((((..........))))))...))).))..)))))...))))))))))).). ( -32.70) >consensus UGGUACGAUUCACUCUGUGACUCAUUGUAAUUGGCAACUUCCGCUGCGCCGGGCGGAAUUAAUGAAAUGUCUUCCCAAAUCGACAUUGAACAGCUAAUUGAGUAAAACAUGAAUUGCCCU .....(((((((...(((.(((((.....((((((...(((((((......))))))).......((((((..........)))))).....)))))))))))...)))))))))).... (-26.22 = -27.42 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:41 2006