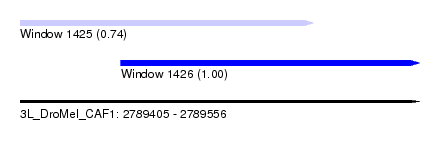
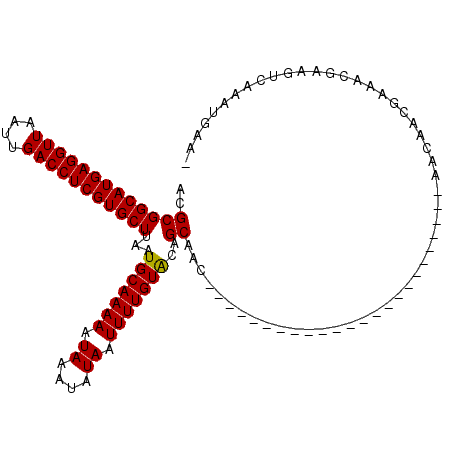
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,789,405 – 2,789,556 |
| Length | 151 |
| Max. P | 0.999271 |

| Location | 2,789,405 – 2,789,516 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 83.01 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -22.45 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.744430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2789405 111 + 23771897 UGGCGCGCUGUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA----U-UU-UGUACAGCA-ACGGCAGCAGC ..((..(((((.(((((.(((((...........))))).))(((((((((((....)))))))))))...(((((((.((....)).----)-))-))))..)))-))))).))... ( -37.60) >DroGri_CAF1 27068 106 + 1 UGU----UAGUGUGUGCUUUGCGUUUGUUGAACUCGCGCUGUGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA---UUUUU-UGUACAGCA-GCA---ACAGC ...----...(((.((((..(((..(.....)..)))((((((((((((((((....)))))))))))....(((((((.........---.))))-)))))))))-)))---))).. ( -34.50) >DroSim_CAF1 24510 111 + 1 UGGCGCGCUGUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA----U-UU-UGUACAGCA-ACGGCAGCAGC ..((..(((((.(((((.(((((...........))))).))(((((((((((....)))))))))))...(((((((.((....)).----)-))-))))..)))-))))).))... ( -37.60) >DroEre_CAF1 36622 102 + 1 UGGCGCGCUGUAUGUGCUUUGCGUUUAUUGAACUUGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA----U-UU-UGUGCGGCA-AC--------- ((.((((((((((..((.(((((...........))))).))(((((((((((....)))))))))))..)))))(((((.......)----)-))-)))))).))-..--------- ( -35.20) >DroMoj_CAF1 28369 112 + 1 UGU----GUGUGAUUGCUUAGCGUUUGUUGAACUCGCCUUGUGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAAGUUUUUUU-UUUACAGCA-GCAGCAGCAGC ...----......(((((..((..((((((.....((.(((.(((((((((((....)))))))))))))).))(((((.((((....)))).)))-))..)))))-)..))))))). ( -31.40) >DroAna_CAF1 24362 103 + 1 UG------AGUGUGUGCUUUGCGUUUAUUGAACUCCCGUCGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA----U-UUUUGAACAGAAAACGGCAG---- .(------(((....))))(((((((.(((............(((((((((((....))))))))))).....(((((((.......)----)-)))))..))).)))).))).---- ( -26.60) >consensus UGG____CUGUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA____U_UU_UGUACAGCA_ACGGCAGCAGC .........((...((((.(((((((...))))..(((....(((((((((((....)))))))))))...)))........................))).)))).))......... (-22.45 = -22.98 + 0.53)

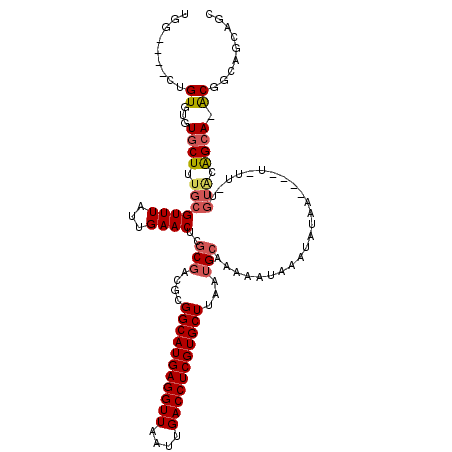
| Location | 2,789,443 – 2,789,556 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.76 |
| Mean single sequence MFE | -27.52 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.35 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.52 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2789443 113 + 23771897 ACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAAUUUUGUACAGCAACGGCAGCAGCAACAACAACAGCGCCGACAACGAAACGAAGUCAAAUGAAA ..(((((((((((((....)))))))))))...(((((((.((....)).)))))))..))...(((.((.............)))))(((..((...))..)))........ ( -30.32) >DroSim_CAF1 24548 110 + 1 ACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAAUUUUGUACAGCAACGGCAGCAGCAACA---ACAACGCCGACAACGAAACGAAGUCAAAUGAAA ..(((((((((((((....)))))))))))...(((((((.((....)).)))))))..))...(((..........---.....)))(((..((...))..)))........ ( -28.16) >DroEre_CAF1 36660 88 + 1 ACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAAUUUUGUGCGGCAAC------------------------AACAACGAGGCGAAGUCAAAUGAA- .((((((((((((((....)))))))))))...(((....(((((......)))))...)))..------------------------.........)))............- ( -25.80) >DroYak_CAF1 35810 88 + 1 ACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAAUUUUGUGCGGCAAC------------------------AACAACGAGGCGAAGUCAAAUGAA- .((((((((((((((....)))))))))))...(((....(((((......)))))...)))..------------------------.........)))............- ( -25.80) >consensus ACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAAUUUUGUACAGCAAC________________________AACAACGAAACGAAGUCAAAUGAA_ ..(((((((((((((....)))))))))))...(((((((.((....)).)))))))..)).................................................... (-21.60 = -21.35 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:37 2006