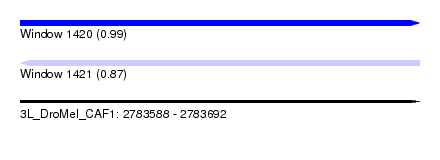
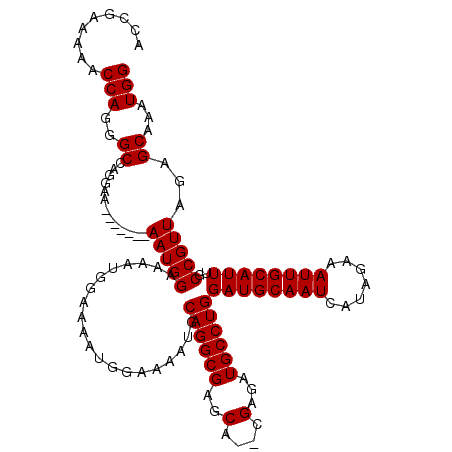
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,783,588 – 2,783,692 |
| Length | 104 |
| Max. P | 0.988639 |

| Location | 2,783,588 – 2,783,692 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -19.26 |
| Energy contribution | -19.30 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2783588 104 + 23771897 CCAUUUGCUCUAACGGCAAAUGCAAUUUCUAUGAUUGCAUCCAGGCAUCUCGCCUGCUCGCCUGCAUUUUCCAUU-------UUCCAUU-------UUCCUGGCCCUGGUUUUUCGGU ((...(((......(((..((((((((.....)))))))).(((((.....)))))...))).)))....(((..-------..(((..-------....)))...)))......)). ( -24.80) >DroSec_CAF1 28956 109 + 1 CCAUUUGCUCUAACGGCAAAUGCAAUUUCUAUUAUUGCAUCCAGGCAUCUCG--UGCUCGCCUGCAUUUUCCAUUUUCCAUUUUCCAUU-------UUCCUGGCCCUGGUUUUUCGGU ((((((((.(....)))))))(((((.......)))))..(((((......(--(((......)))).................(((..-------....))).)))))......)). ( -23.00) >DroSim_CAF1 18404 102 + 1 CCAUUUGCUCUAACGGCAAAUGCAAUUUCUAUGAUUGCAUCCAGGCAUCUCG--UGCUCGCCUGCAUUUUCCAUU-------UUCCAUU-------UUCCUGGCCCUGGUUUUUCGGU ((((((((.(....)))))))((((((.....))))))..(((((......(--(((......))))........-------..(((..-------....))).)))))......)). ( -24.60) >DroEre_CAF1 30549 116 + 1 CCAUUUGCUCUAACGGCAAAUGCAAUUUCUAUGAUUGCAUCCAGGCAUCUCG--UGCUCGCCUGCAUUUCCCAUUUUCCAUUUUCCAUUUUCCCUUUGCCUGGCCCUGGUUUUUCGUU ..((((((.(....)))))))((((((.....))))))..((((((((...)--)))..(((.(((..............................)))..))).))))......... ( -24.41) >DroYak_CAF1 29682 109 + 1 CCAUUUGCUCUAACGGCAAAUGCAAUUUCUAUGAUUGCAUCCAGGCAUCUCG--UGCCCGCCUGCAUU-------UUCCAUUUUCCAUUUUCCCUUUUCCUGGCUCUGGUUUUUCGGU ((((((((.(....)))))))((((((.....))))))..((((((((...)--)))).(((......-------..........................)))..)))......)). ( -22.15) >consensus CCAUUUGCUCUAACGGCAAAUGCAAUUUCUAUGAUUGCAUCCAGGCAUCUCG__UGCUCGCCUGCAUUUUCCAUUUUCCAUUUUCCAUU_______UUCCUGGCCCUGGUUUUUCGGU ..(((((((.....)))))))((((((.....))))))..(((((.........(((......)))..................(((.............))).)))))......... (-19.26 = -19.30 + 0.04)

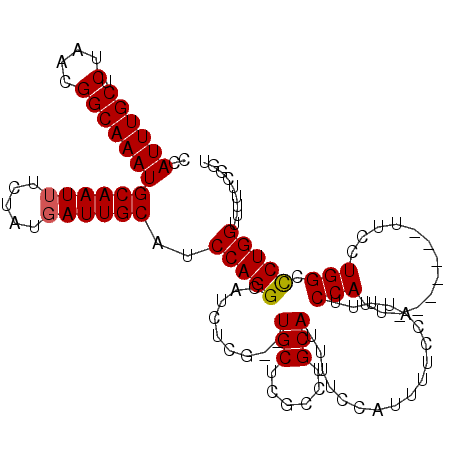
| Location | 2,783,588 – 2,783,692 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 87.46 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -20.07 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2783588 104 - 23771897 ACCGAAAAACCAGGGCCAGGAA-------AAUGGAA-------AAUGGAAAAUGCAGGCGAGCAGGCGAGAUGCCUGGAUGCAAUCAUAGAAAUUGCAUUUGCCGUUAGAGCAAAUGG .(((.....(((...(((....-------..)))..-------..)))....(((.(((((.(((((.....))))).(((((((.......))))))))))))......)))..))) ( -28.80) >DroSec_CAF1 28956 109 - 1 ACCGAAAAACCAGGGCCAGGAA-------AAUGGAAAAUGGAAAAUGGAAAAUGCAGGCGAGCA--CGAGAUGCCUGGAUGCAAUAAUAGAAAUUGCAUUUGCCGUUAGAGCAAAUGG .(((.....(((...(((....-------..)))....)))..(((((......((((((..(.--...).))))))((((((((.......))))))))..)))))........))) ( -26.20) >DroSim_CAF1 18404 102 - 1 ACCGAAAAACCAGGGCCAGGAA-------AAUGGAA-------AAUGGAAAAUGCAGGCGAGCA--CGAGAUGCCUGGAUGCAAUCAUAGAAAUUGCAUUUGCCGUUAGAGCAAAUGG .((......(((...(((....-------..)))..-------..)))...((.((((((..(.--...).)))))).))(((((.......)))))(((((((....).)))))))) ( -23.90) >DroEre_CAF1 30549 116 - 1 AACGAAAAACCAGGGCCAGGCAAAGGGAAAAUGGAAAAUGGAAAAUGGGAAAUGCAGGCGAGCA--CGAGAUGCCUGGAUGCAAUCAUAGAAAUUGCAUUUGCCGUUAGAGCAAAUGG .........(((...(((.............)))....)))...((((...((.((((((..(.--...).)))))).))....))))........((((((((....).))))))). ( -24.82) >DroYak_CAF1 29682 109 - 1 ACCGAAAAACCAGAGCCAGGAAAAGGGAAAAUGGAAAAUGGAA-------AAUGCAGGCGGGCA--CGAGAUGCCUGGAUGCAAUCAUAGAAAUUGCAUUUGCCGUUAGAGCAAAUGG .(((.....(((...((........))....)))....)))..-------..((((..((((((--.....))))))..)))).............((((((((....).))))))). ( -28.60) >consensus ACCGAAAAACCAGGGCCAGGAA_______AAUGGAAAAUGGAAAAUGGAAAAUGCAGGCGAGCA__CGAGAUGCCUGGAUGCAAUCAUAGAAAUUGCAUUUGCCGUUAGAGCAAAUGG .........(((..((.............(((((....................((((((..(....)...))))))((((((((.......))))))))..)))))...))...))) (-20.07 = -20.07 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:32 2006