| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,777,165 – 2,777,255 |
| Length | 90 |
| Max. P | 0.595662 |

| Location | 2,777,165 – 2,777,255 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.03 |
| Mean single sequence MFE | -37.62 |
| Consensus MFE | -18.62 |
| Energy contribution | -17.57 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
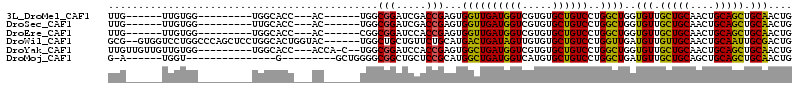
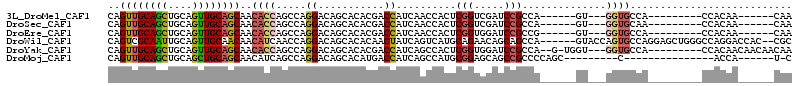
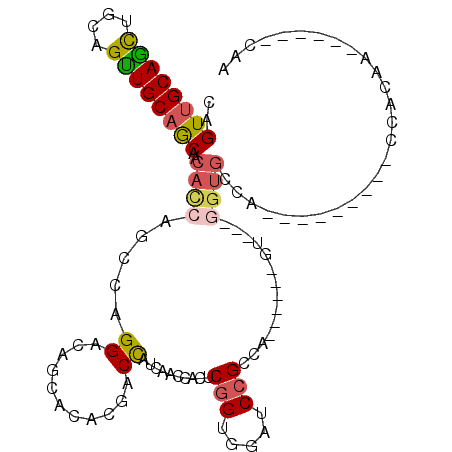
>3L_DroMel_CAF1 2777165 90 + 23771897 UUG------UUGUGG---------UGGCACC---AC------UGGCGGAUCGACCGAGUGGUUGAUGGUCGUGUGCUGUCCUGGCUGGUGUUGCUGCAACUGCAGCUGCAACUG ..(------(((..(---------..(((((---((------((((((..((((((.........))))))....)))))..)).))))))..)..))))(((....))).... ( -37.60) >DroSec_CAF1 22480 90 + 1 UUG------UUGUGG---------UUGCACC---AC------UGGCGGAUCGACCGAGUGGUUGAUGGUCGUGUGCUGUCCUGGCUGGUGUUGCUGCAACUGCAGCUGCAACUG ...------....((---------(((((.(---((------..((..(((((((....)))))))((.((.....)).))..))..)))..(((((....)))))))))))). ( -36.10) >DroEre_CAF1 24105 90 + 1 UUG------UUGUGG---------UGGCACC---AC------CGGCGGAUCCACCGAGUGGUUGAUGGUCGUGUGCUGUCCUGGCUGGUGUUGCUGCAACUGCAGCUGCAACUG ..(------(((..(---------..(((((---((------(((.((...((((((.((.....)).))).)))...)))))).))))))..)..))))(((....))).... ( -37.60) >DroWil_CAF1 12201 106 + 1 GCG--GUGGUCCUGGCCCAGCUCCUGGCACUGGUAC------UGGCUGCUGUUCUGCAUGACUGAUAGUUGUGUGCUGUCCUGGUUGAUGUUGUUGCAACUGCAAUUGCGACUG (((--(..(((..(..((((.........))))..)------.)))..))))...((((((((((((((.....))))))..)))).)))).(((((((......))))))).. ( -32.50) >DroYak_CAF1 21743 99 + 1 UUGUUGUUGUUGUGG---------UGGCACC---ACCA-C--UGGCGGAUCCACCGAGUGGCUGAUGGUCGUGUGCUGUCCUGGCUGGUGUUGCUGCAACUGCAGCUGCAACUG ..(((((.((((..(---------..(((((---((((-.--.(((((...((((((.((.....)).))).)))))))).))).))))))..)..)))).)))))........ ( -42.60) >DroMoj_CAF1 12798 83 + 1 G-A------UGGU---------------G---------GCUGGGGCGGCUGCUCCGCAUGGCUGAUGGUCAUGUGCUGUCCUGGCUGAUGUUGCUGCAGCUGCAGCUGCAACUG .-.------.(((---------------(---------((..(((((((......(((((((.....))))))))))))))..)))..(((.(((((....))))).)))))). ( -39.30) >consensus UUG______UUGUGG_________UGGCACC___AC______UGGCGGAUCCACCGAGUGGCUGAUGGUCGUGUGCUGUCCUGGCUGGUGUUGCUGCAACUGCAGCUGCAACUG .............................................(((.....)))...((((((((((.....))))))..))))..(((.(((((....))))).))).... (-18.62 = -17.57 + -1.05)
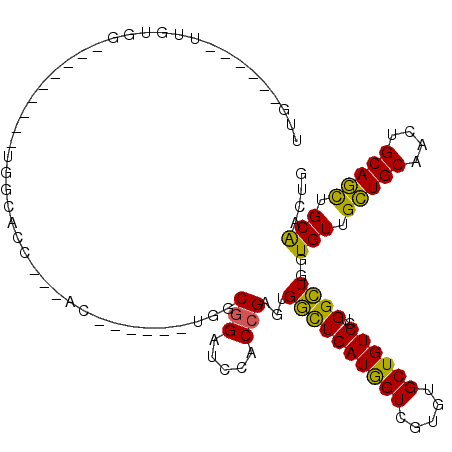
| Location | 2,777,165 – 2,777,255 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 75.03 |
| Mean single sequence MFE | -27.30 |
| Consensus MFE | -14.09 |
| Energy contribution | -14.45 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.595662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2777165 90 - 23771897 CAGUUGCAGCUGCAGUUGCAGCAACACCAGCCAGGACAGCACACGACCAUCAACCACUCGGUCGAUCCGCCA------GU---GGUGCCA---------CCACAA------CAA ..((((..(((((....)))))..(((((((.......))...(((((...........)))))........------.)---))))...---------...)))------).. ( -26.20) >DroSec_CAF1 22480 90 - 1 CAGUUGCAGCUGCAGUUGCAGCAACACCAGCCAGGACAGCACACGACCAUCAACCACUCGGUCGAUCCGCCA------GU---GGUGCAA---------CCACAA------CAA ..(((((.(((((....)))))....((.....)).................((((((.(((......))))------))---)))))))---------).....------... ( -27.40) >DroEre_CAF1 24105 90 - 1 CAGUUGCAGCUGCAGUUGCAGCAACACCAGCCAGGACAGCACACGACCAUCAACCACUCGGUGGAUCCGCCG------GU---GGUGCCA---------CCACAA------CAA ..((((..(((((....)))))...........((...((....(.....).(((((.(((((....)))))------))---)))))..---------)).)))------).. ( -31.10) >DroWil_CAF1 12201 106 - 1 CAGUCGCAAUUGCAGUUGCAACAACAUCAACCAGGACAGCACACAACUAUCAGUCAUGCAGAACAGCAGCCA------GUACCAGUGCCAGGAGCUGGGCCAGGACCAC--CGC ..((.(((((....))))).))........((.((.((((................(((......))).((.------(((....)))..)).))))..)).)).....--... ( -21.50) >DroYak_CAF1 21743 99 - 1 CAGUUGCAGCUGCAGUUGCAGCAACACCAGCCAGGACAGCACACGACCAUCAGCCACUCGGUGGAUCCGCCA--G-UGGU---GGUGCCA---------CCACAACAACAACAA ..((((..(((((....)))))...........((...((((..(.....).((((((.((((....)))))--)-))))---.))))..---------)).))))........ ( -34.20) >DroMoj_CAF1 12798 83 - 1 CAGUUGCAGCUGCAGCUGCAGCAACAUCAGCCAGGACAGCACAUGACCAUCAGCCAUGCGGAGCAGCCGCCCCAGC---------C---------------ACCA------U-C ..((((((((....))))))))...........((...((.((((.........)))).((.((....)).)).))---------.---------------.)).------.-. ( -23.40) >consensus CAGUUGCAGCUGCAGUUGCAGCAACACCAGCCAGGACAGCACACGACCAUCAACCACUCGGUGGAUCCGCCA______GU___GGUGCCA_________CCACAA______CAA ..((((((((....))))))))..((((.....((...........))..........(((.....)))..............))))........................... (-14.09 = -14.45 + 0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:25 2006