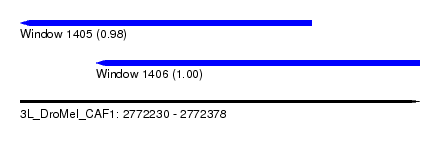
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,772,230 – 2,772,378 |
| Length | 148 |
| Max. P | 0.999469 |

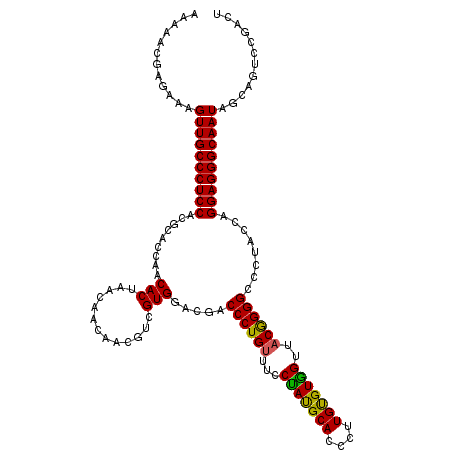
| Location | 2,772,230 – 2,772,338 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.72 |
| Mean single sequence MFE | -45.88 |
| Consensus MFE | -42.15 |
| Energy contribution | -41.43 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2772230 108 - 23771897 CCACGUCGUGGACGACCCUGUUCCCCACGCAUCCGUGCGUGGUUUCGGGGAAAUACCCGGAGGGCAAUAGCAGUCCGACCUCGUCCACGAACAACGCGCCC------A---ACGGUG .....(((((((((((((((....(((((((....)))))))...))))).......((((..((....))..))))...))))))))))......((((.------.---..)))) ( -49.20) >DroVir_CAF1 7671 108 - 1 CAACGUCGUGGACGACCCUGUUUCCUAUGCACCCAUGUGUGGUUACGGGGCCCUAUCAGGAGGGCAAUAGCAGUCCAACCUCGUCCACGAAUGGCAGCGCC------A---ACGCCA .....((((((((((((((((..((.(((((....)))))))..))))))........(((..((....))..)))....)))))))))).((((...)))------)---...... ( -42.80) >DroWil_CAF1 7253 98 - 1 CCACGUCGUGGACGACCCUGUUUCCUAUGCACCCUUGUGUGGUUACAGGGCCCUACCAGGAGGGCAAUAGCAGUCCAACUUCGUCCACGAAUGGCCA-------------------G (((..((((((((((((((((..((.(((((....)))))))..))))))........(((..((....))..)))....)))))))))).)))...-------------------. ( -42.20) >DroMoj_CAF1 7767 111 - 1 CAACGUCGUGGACGACCCUGUUCCCUAUGCACCCAUGCGUAGUUACGGGGCCCUACCAGGAGGGCAAUAGCAGUCCAACAUCGUCCACGAAUGGCAGCGCC------ACCAAUGGCA ...((((((((((((((((((...(((((((....)))))))..))))))........(((..((....))..)))....))))))))).))).....(((------(....)))). ( -47.30) >DroAna_CAF1 8202 114 - 1 CCACGUCGUGGACGACCCUGUUCCCCAUGCAUCCGUGCGUGGUUACGGGGACCUAUCCGGAGGGCAAUAGCAGUCCGACCUCGUCCACGAACAACAACAGCAACAGCA---ACGCUG .....((((((((((((((((...(((((((....)))))))..)))))).......((((..((....))..))))...)))))))))).......((((.......---..)))) ( -49.80) >DroPer_CAF1 20175 108 - 1 CAACAUCGUGGACGACCCUGUUUCCUAUGCAUCCUUGUGUGGUUACGGGGCCCUAUCAGGAGGGCAAUAGCAGUCCGACUUCGUCCACGAAUGGCAGCGCC------A---ACAACA .....((((((((((((((((..((.(((((....)))))))..))))))........(((..((....))..)))....)))))))))).((((...)))------)---...... ( -44.00) >consensus CAACGUCGUGGACGACCCUGUUCCCUAUGCACCCAUGCGUGGUUACGGGGCCCUACCAGGAGGGCAAUAGCAGUCCAACCUCGUCCACGAAUGGCAGCGCC______A___ACGCCA .....((((((((((((((((...(((((((....)))))))..))))))........(((..((....))..)))....))))))))))........................... (-42.15 = -41.43 + -0.72)

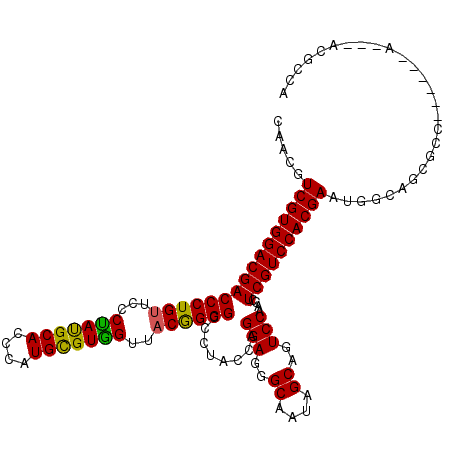
| Location | 2,772,258 – 2,772,378 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -37.43 |
| Energy contribution | -36.82 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.63 |
| SVM RNA-class probability | 0.999469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2772258 120 - 23771897 AAAAACGAGAAGGUUGCCCUCCACGCACCAACACUGACGACCACGUCGUGGACGACCCUGUUCCCCACGCAUCCGUGCGUGGUUUCGGGGAAAUACCCGGAGGGCAAUAGCAGUCCGACC .....((.((..((((((((((.........(((.((((....))))))).....(((((....(((((((....)))))))...)))))........)))))))))).....))))... ( -50.00) >DroPse_CAF1 20496 120 - 1 AAAAACGAGAAAGUUGCCCUCCACGCACCAACACUAACAACAACAUCGUGGACGACCCUGUUUCCUAUGCAUCCUUGUGUGGUUACGGGGCCCUAUCAGGAGGGCAAUAGCAGUCCGACU .....((.((..((((((((((.......................(((....)))((((((..((.(((((....)))))))..))))))........)))))))))).....))))... ( -38.50) >DroGri_CAF1 7520 120 - 1 AAAAACGAGAAAGUUGCCCUCCACGCACCAACACUAACAACAACGUCGUGGACAACCCUGUUUCCUAUGCACCCAUGUGUGGUUACGGGGCCCUAUCAGGAGGGCAAUAGCAGUCCGACA .....((.((..((((((((((.....(((..((..........))..)))....((((((..((.(((((....)))))))..))))))........)))))))))).....))))... ( -37.70) >DroWil_CAF1 7271 120 - 1 AAAAACGAGAAAGUUGCCCUCCACGCACCAACACUAACAACCACGUCGUGGACGACCCUGUUUCCUAUGCACCCUUGUGUGGUUACAGGGCCCUACCAGGAGGGCAAUAGCAGUCCAACU ............((((((((((..................((((...))))....((((((..((.(((((....)))))))..))))))........))))))))))............ ( -39.00) >DroMoj_CAF1 7798 120 - 1 AAAAACGAGAAAGUUGCCCUCCACGCACCAACACUAACAACAACGUCGUGGACGACCCUGUUCCCUAUGCACCCAUGCGUAGUUACGGGGCCCUACCAGGAGGGCAAUAGCAGUCCAACA ............((((((((((.....................((((...)))).((((((...(((((((....)))))))..))))))........))))))))))............ ( -40.00) >DroPer_CAF1 20203 120 - 1 AAAAACGAGAAAGUUGCCCUCCACGCACCAACACUAACAACAACAUCGUGGACGACCCUGUUUCCUAUGCAUCCUUGUGUGGUUACGGGGCCCUAUCAGGAGGGCAAUAGCAGUCCGACU .....((.((..((((((((((.......................(((....)))((((((..((.(((((....)))))))..))))))........)))))))))).....))))... ( -38.50) >consensus AAAAACGAGAAAGUUGCCCUCCACGCACCAACACUAACAACAACGUCGUGGACGACCCUGUUUCCUAUGCACCCUUGUGUGGUUACGGGGCCCUACCAGGAGGGCAAUAGCAGUCCGACU ............((((((((((.........(((.............))).....((((((...(((((((....)))))))..))))))........))))))))))............ (-37.43 = -36.82 + -0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:17 2006