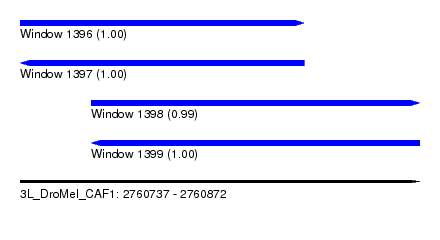
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,760,737 – 2,760,872 |
| Length | 135 |
| Max. P | 0.999996 |

| Location | 2,760,737 – 2,760,833 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.96 |
| Mean single sequence MFE | -35.37 |
| Consensus MFE | -27.41 |
| Energy contribution | -26.53 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.77 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998368 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2760737 96 + 23771897 ----------------GUUUAGACGAAACCUGUUGCUCAGUACGAGCGCCAUGGAAAGUAGUAUUUACUUUUCCAUGGCGGAGCCAACGGCUUCUGCUCCACCGGCGCCAC-C------- ----------------..............((.((((..((..((((((((((((((((((...)))).))))))))))((((((...)))))).)))).)).)))).)).-.------- ( -34.30) >DroSim_CAF1 116358 96 + 1 ----------------GUUCAAAAGAAACCUGCUGCUCAGUACGAGUGCCAUGGAGAGCAGCAUCUACUUUUCCAUGGCGGAGCCAACGGUUUCUGCUCCACCGGCGCCAC-C------- ----------------.......(((((((.(((((((.....))))(((((((((((..........)))))))))))..)))....)))))))((.((...)).))...-.------- ( -31.20) >DroAna_CAF1 114227 120 + 1 AGGAGGACGAAGACCAGUUCCGCCGCAAUCUGCUGCUGAGCACCAGCACCAUGGAGAGCAGCAUAUACUUCUCCAUGGCAGAGCCGACAACGCCUGCUCCGGCAGCUGCUCCCAGCCCGA .((((...........((((.((.((.....)).)).))))..((((.((((((((((..........))))))))))....((((.............)))).))))))))........ ( -40.62) >consensus ________________GUUCAGACGAAACCUGCUGCUCAGUACGAGCGCCAUGGAGAGCAGCAUAUACUUUUCCAUGGCGGAGCCAACGGCUUCUGCUCCACCGGCGCCAC_C_______ ...............................(((((((.....))))(((((((((((..........)))))))))))(((((...........)))))...))).............. (-27.41 = -26.53 + -0.88)

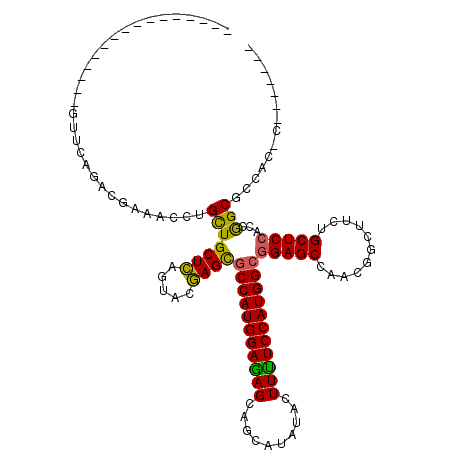
| Location | 2,760,737 – 2,760,833 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.96 |
| Mean single sequence MFE | -44.10 |
| Consensus MFE | -33.14 |
| Energy contribution | -31.60 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.75 |
| SVM decision value | 6.09 |
| SVM RNA-class probability | 0.999996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2760737 96 - 23771897 -------G-GUGGCGCCGGUGGAGCAGAAGCCGUUGGCUCCGCCAUGGAAAAGUAAAUACUACUUUCCAUGGCGCUCGUACUGAGCAACAGGUUUCGUCUAAAC---------------- -------.-((.((..(((((((((.(.((((...)))).)((((((((((.(((.....))))))))))))))))).))))).)).))...............---------------- ( -38.00) >DroSim_CAF1 116358 96 - 1 -------G-GUGGCGCCGGUGGAGCAGAAACCGUUGGCUCCGCCAUGGAAAAGUAGAUGCUGCUCUCCAUGGCACUCGUACUGAGCAGCAGGUUUCUUUUGAAC---------------- -------(-((((.(((..(((........)))..))).)))))....(((((....((((((((...(((.....)))...)))))))).....)))))....---------------- ( -38.30) >DroAna_CAF1 114227 120 - 1 UCGGGCUGGGAGCAGCUGCCGGAGCAGGCGUUGUCGGCUCUGCCAUGGAGAAGUAUAUGCUGCUCUCCAUGGUGCUGGUGCUCAGCAGCAGAUUGCGGCGGAACUGGUCUUCGUCCUCCU ..((((..(((.(((((((((((((.(((...))).)))).((((((((((.(((.....))))))))))))).(((.(((...))).)))....))))))..))).)))..)))).... ( -56.00) >consensus _______G_GUGGCGCCGGUGGAGCAGAAGCCGUUGGCUCCGCCAUGGAAAAGUAAAUGCUGCUCUCCAUGGCGCUCGUACUGAGCAGCAGGUUUCGUCUGAAC________________ ...........((.(((.(((((((.((.....)).)))))((((((((((.(((.....)))))))))))))((((.....)))).)).))).))........................ (-33.14 = -31.60 + -1.54)

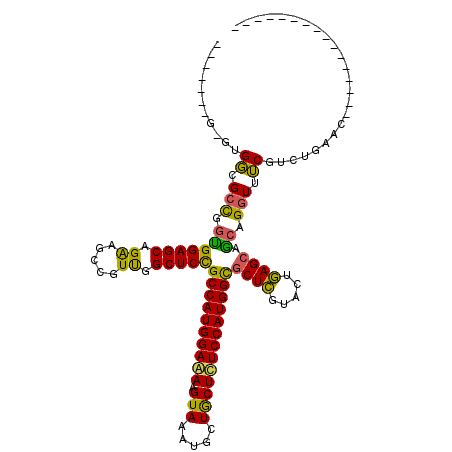
| Location | 2,760,761 – 2,760,872 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -28.47 |
| Energy contribution | -28.37 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2760761 111 + 23771897 UACGAGCGCCAUGGAAAGUAGUAUUUACUUUUCCAUGGCGGAGCCAACGGCUUCUGCUCCACCGGCGCCAC-C--------GGUGCGUAAUGAAGCCUUCUCCUUUGAGUUGUUUGUAGA (((((((((((((((((((((...)))).))))))))))((((.....((((((....(((((((.....)-)--------)))).)....))))))..))))........))))))).. ( -49.50) >DroSim_CAF1 116382 111 + 1 UACGAGUGCCAUGGAGAGCAGCAUCUACUUUUCCAUGGCGGAGCCAACGGUUUCUGCUCCACCGGCGCCAC-C--------AGUGCGGAAUGAAGCCUUCUCCUUUGAGUUGUUUGUGGA ((((((.(((((((((((..........)))))))))))((((.....((((((..((.(((.((.....)-)--------.))).))...))))))..)))).........)))))).. ( -36.60) >DroAna_CAF1 114267 120 + 1 CACCAGCACCAUGGAGAGCAGCAUAUACUUCUCCAUGGCAGAGCCGACAACGCCUGCUCCGGCAGCUGCUCCCAGCCCGAAGGUGAGCAGCGAAGCCUUCUCCUUCGAGCUGUUCGUCGA .....((.((((((((((..........))))))))))....))((((((((...((((.....(((((((((........)).)))))))((((.......)))))))))))).)))). ( -44.40) >consensus UACGAGCGCCAUGGAGAGCAGCAUAUACUUUUCCAUGGCGGAGCCAACGGCUUCUGCUCCACCGGCGCCAC_C________GGUGCGCAAUGAAGCCUUCUCCUUUGAGUUGUUUGUAGA ((((((((((((((((((..........)))))))))))..((((((.((.....(((.((...(((.(((...........))))))..)).))).....)).))).)))))))))).. (-28.47 = -28.37 + -0.10)

| Location | 2,760,761 – 2,760,872 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.35 |
| Mean single sequence MFE | -46.57 |
| Consensus MFE | -33.12 |
| Energy contribution | -31.59 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.75 |
| SVM RNA-class probability | 0.999583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2760761 111 - 23771897 UCUACAAACAACUCAAAGGAGAAGGCUUCAUUACGCACC--------G-GUGGCGCCGGUGGAGCAGAAGCCGUUGGCUCCGCCAUGGAAAAGUAAAUACUACUUUCCAUGGCGCUCGUA ..(((............((((..((((((....(.((((--------(-(.....)))))))....)))))).....))))((((((((((.(((.....)))))))))))))....))) ( -46.10) >DroSim_CAF1 116382 111 - 1 UCCACAAACAACUCAAAGGAGAAGGCUUCAUUCCGCACU--------G-GUGGCGCCGGUGGAGCAGAAACCGUUGGCUCCGCCAUGGAAAAGUAGAUGCUGCUCUCCAUGGCACUCGUA .......((........((((..((.(((.((((..(((--------(-(.....)))))))))..))).)).....))))((((((((..(((((...))))).))))))))....)). ( -37.40) >DroAna_CAF1 114267 120 - 1 UCGACGAACAGCUCGAAGGAGAAGGCUUCGCUGCUCACCUUCGGGCUGGGAGCAGCUGCCGGAGCAGGCGUUGUCGGCUCUGCCAUGGAGAAGUAUAUGCUGCUCUCCAUGGUGCUGGUG ........(((((((((((.((.(((...)))..)).)))))))))))..(.((((....(((((.(((...))).)))))((((((((((.(((.....))))))))))))))))).). ( -56.20) >consensus UCCACAAACAACUCAAAGGAGAAGGCUUCAUUACGCACC________G_GUGGCGCCGGUGGAGCAGAAGCCGUUGGCUCCGCCAUGGAAAAGUAAAUGCUGCUCUCCAUGGCGCUCGUA ...........(((....)))..(((......((((.............)))).)))...(((((.((.....)).)))))((((((((((.(((.....)))))))))))))....... (-33.12 = -31.59 + -1.54)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:10 2006