
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,730,038 – 2,730,131 |
| Length | 93 |
| Max. P | 0.999786 |

| Location | 2,730,038 – 2,730,131 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -23.58 |
| Consensus MFE | -20.50 |
| Energy contribution | -20.54 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.87 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2730038 93 + 23771897 ACAGCGGUUUGAUUGAAACGCUGAUUGUUGGAGCGUAGCAAAUAUUUACAAAUGUAAACCAGGGGGAAAAAAAAAACAACUAAGCCGCACAAA ...((((((((.(((..(((((.........)))))........(((((....)))))((....))..........))).))))))))..... ( -21.30) >DroSec_CAF1 89963 89 + 1 ACAGCGGUUUGAUUGAAACGCUGAUUGUUGGAGCGUAGCAAAUAUUUACAAAUGUAAACCAGUGCGGAA----AAACAACUAAGCCGCACAAA ...((((((((.(((..(((((.........))))).(((....(((((....)))))....)))....----...))).))))))))..... ( -25.50) >DroSim_CAF1 88318 89 + 1 ACAGCGGUUUGAUUGAAACGCUGAUUGUUGGAGCGUAGCAAAUAUUUACAAAUGUAAACCAGUGCGGAA----AAACAACUAAGCCGCACAAA ...((((((((.(((..(((((.........))))).(((....(((((....)))))....)))....----...))).))))))))..... ( -25.50) >DroEre_CAF1 98534 88 + 1 ACAGCGGUUUGAUUGAAACGCUGAUUGUUGGAGCGUAGCAAAUAUUUACAAUUGUAAACCAGGG-UAAA----AAUCAACUAAGCCGCACAAA ...((((((((.((((.((.(((.((((((.....))))))...(((((....))))).))).)-)...----..)))).))))))))..... ( -23.40) >DroYak_CAF1 91905 88 + 1 ACAGCGGUUUGAUUGAAACGCUGAUUGUUGGAGCGUAGCAAAUAUUUACAAAUGUAAACCAGGC-UAAA----AAUCAACUAUACCGCACAAA ...(((((.((.((((.(((((.........)))))(((.....(((((....)))))....))-)...----..)))).)).)))))..... ( -22.20) >consensus ACAGCGGUUUGAUUGAAACGCUGAUUGUUGGAGCGUAGCAAAUAUUUACAAAUGUAAACCAGGG_GAAA____AAACAACUAAGCCGCACAAA ...((((((((.(((..(((((.........)))))..)))...(((((....)))))......................))))))))..... (-20.50 = -20.54 + 0.04)

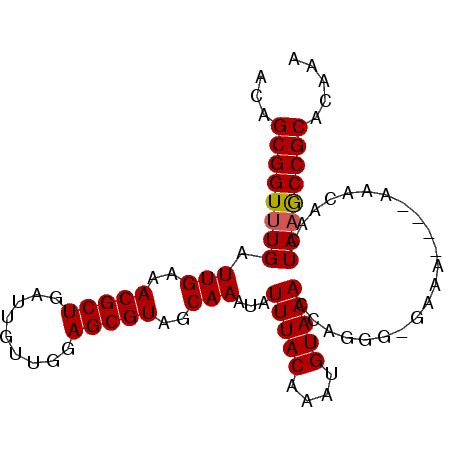
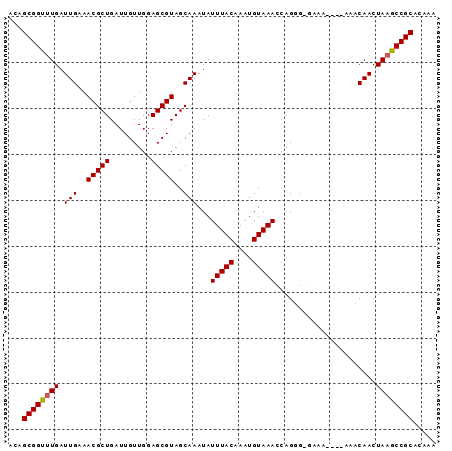
| Location | 2,730,038 – 2,730,131 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 92.93 |
| Mean single sequence MFE | -19.44 |
| Consensus MFE | -16.14 |
| Energy contribution | -16.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916410 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2730038 93 - 23771897 UUUGUGCGGCUUAGUUGUUUUUUUUUUCCCCCUGGUUUACAUUUGUAAAUAUUUGCUACGCUCCAACAAUCAGCGUUUCAAUCAAACCGCUGU .....((((.........................((((((....))))))..(((..(((((.........)))))..))).....))))... ( -16.10) >DroSec_CAF1 89963 89 - 1 UUUGUGCGGCUUAGUUGUUU----UUCCGCACUGGUUUACAUUUGUAAAUAUUUGCUACGCUCCAACAAUCAGCGUUUCAAUCAAACCGCUGU .....((((.((.((((...----....(((...((((((....))))))...))).(((((.........)))))..)))).)).))))... ( -20.10) >DroSim_CAF1 88318 89 - 1 UUUGUGCGGCUUAGUUGUUU----UUCCGCACUGGUUUACAUUUGUAAAUAUUUGCUACGCUCCAACAAUCAGCGUUUCAAUCAAACCGCUGU .....((((.((.((((...----....(((...((((((....))))))...))).(((((.........)))))..)))).)).))))... ( -20.10) >DroEre_CAF1 98534 88 - 1 UUUGUGCGGCUUAGUUGAUU----UUUA-CCCUGGUUUACAAUUGUAAAUAUUUGCUACGCUCCAACAAUCAGCGUUUCAAUCAAACCGCUGU .((((..(((.((((.(((.----((((-(..((.....))...))))).))).)))).)))...)))).((((((((.....))).))))). ( -19.00) >DroYak_CAF1 91905 88 - 1 UUUGUGCGGUAUAGUUGAUU----UUUA-GCCUGGUUUACAUUUGUAAAUAUUUGCUACGCUCCAACAAUCAGCGUUUCAAUCAAACCGCUGU .....(((((.(.(((((..----..((-((...((((((....))))))....))))((((.........))))..))))).).)))))... ( -21.90) >consensus UUUGUGCGGCUUAGUUGUUU____UUUC_CCCUGGUUUACAUUUGUAAAUAUUUGCUACGCUCCAACAAUCAGCGUUUCAAUCAAACCGCUGU .....((((.........................((((((....))))))..(((..(((((.........)))))..))).....))))... (-16.14 = -16.14 + 0.00)

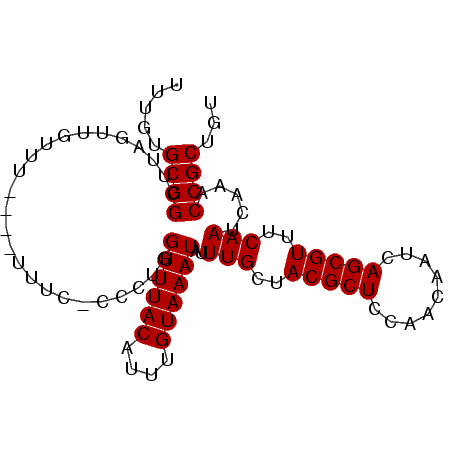
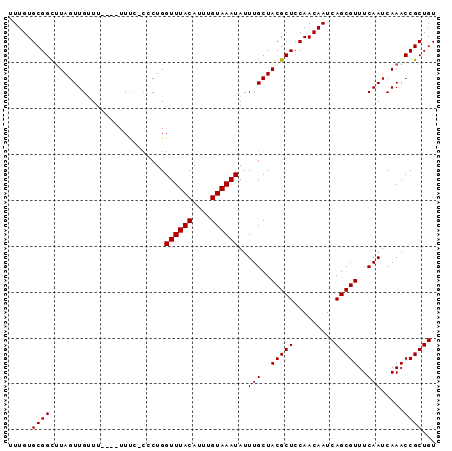
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:54:00 2006