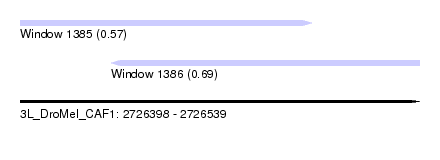
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,726,398 – 2,726,539 |
| Length | 141 |
| Max. P | 0.691211 |

| Location | 2,726,398 – 2,726,501 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.99 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -18.18 |
| Energy contribution | -18.47 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2726398 103 + 23771897 AUCGUAGAUAUAU---UUGGCACAGCGAA-----AGCUGGCGCU-UUUGGAUCUGCAGUUUGUGCUGUUGUUCAGUUUUUG-GUAAUCUGUCUGAUUAUUUAACUUGCGCUAA .............---......((((...-----.))))((((.-..(((((..(((((....))))).)))))(((...(-((((((.....))))))).)))..))))... ( -30.20) >DroSec_CAF1 86546 103 + 1 AUCGUAGAUAUAU---UUGGCACACCGAA-----AGCUGGCGCU-UUUGAAUCUGCAGUUUGUGCUGUUGUUCAGUUUUUG-GUAAUCUGUCUGAUUAUUUAACUUGCGCUAA ............(---((((....)))))-----...((((((.-..(((((..(((((....))))).)))))(((...(-((((((.....))))))).)))..)))))). ( -27.00) >DroSim_CAF1 84759 103 + 1 AUCGUAGAUAUAU---UUGGCACCGCGAA-----AGCUGGCGCU-UUUGAAUCUGCAGUUUGUGCUGUUGUUCAGUUUUUG-GUAAUCUGUCUGAUUAUUUAACUUGCGCUAA ...((((((.((.---..(((.((((...-----.)).)).)))-..)).)))))).....((((........((((...(-((((((.....))))))).)))).))))... ( -28.50) >DroEre_CAF1 94845 101 + 1 AUCGUAGGUAUAU---UUGGCACAUU--U-----CGCUGGCGCU-UUUGGAUCUGCAGUUUGUGCUGUUGCUCAGUUUUUG-GUAAUCUGUCUGAUUAUUUAACUUGCGCUAA .............---..(((.....--.-----.)))(((((.-....((.(.(((((....))))).).))((((...(-((((((.....))))))).)))).))))).. ( -22.50) >DroYak_CAF1 88245 101 + 1 AUCGUUGGUAUAU---UUGGCACUUU--U-----CGCUGGCGCU-UUUGGAUCUGCAGUUUGUGCUGUUGCUCAGUUUUUG-GUAAUCUGUCUGAUUAUUUAACUUGCGCUAA ...((..(.....---)..)).....--.-----...((((((.-....((.(.(((((....))))).).))((((...(-((((((.....))))))).)))).)))))). ( -24.90) >DroAna_CAF1 84813 109 + 1 UACGUAUCUAUAUACUUUCGGGCAGCGGACUAUGCGCUUUUGCUUUUUGGAUUUUUAGUUUCUGG----GUCCAACGUAUGCAUAAUCUGUCUGAUUAUUUAUCUUGCGCUAA ..((((...(((.....((((((((.....(((((((....))...((((((((.........))----)))))).....)))))..)))))))).....)))..)))).... ( -25.40) >consensus AUCGUAGAUAUAU___UUGGCACAGCGAA_____AGCUGGCGCU_UUUGGAUCUGCAGUUUGUGCUGUUGUUCAGUUUUUG_GUAAUCUGUCUGAUUAUUUAACUUGCGCUAA .....................................((((((...((((((..(((((....))))).))))))...(((.((((((.....)))))).)))...)))))). (-18.18 = -18.47 + 0.28)

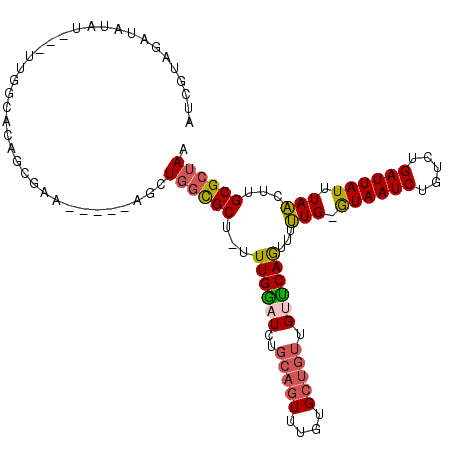
| Location | 2,726,430 – 2,726,539 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.86 |
| Mean single sequence MFE | -15.78 |
| Consensus MFE | -8.05 |
| Energy contribution | -8.38 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.691211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2726430 109 - 23771897 AUAAAAAUUGCCGCAAAAUAUUUUCUAAAUAUUUGUAUUUAGCGCAAGUUAAAUAAUCAGACAGAUUAC-CAAAAACUGAACAACAGCACAAA-CUGCAGA---UCCAAA-AGCG ...........(((.(((((((.....))))))).((((((((....))))))))(((...(((.((..-...)).))).......(((....-.))).))---).....-.))) ( -16.30) >DroVir_CAF1 103639 107 - 1 AUAAAAAUCGCCACAAAAUA-UUUCUAAAUAUUUGUAUUUAGCGCAAGUUAAAUAAUCAGACAGAUUAU-GCAUACGUUAUAAAAAUAACAAAAAGCCAAA---CCGAAA-AA-- .......(((.....(((((-((....)))))))(((((((((....)))))((((((.....))))))-..))))(((((....)))))...........---.)))..-..-- ( -15.10) >DroSec_CAF1 86578 91 - 1 ------------------UUUUUUCUAAAUAUUUGUAUUUAGCGCAAGUUAAAUAAUCAGACAGAUUAC-CAAAAACUGAACAACAGCACAAA-CUGCAGA---UUCAAA-AGCG ------------------.....(((......(((((((((((....)))))))..((((.........-......))))))))..(((....-.))))))---......-.... ( -12.66) >DroSim_CAF1 84791 109 - 1 AUAAAAAUUGCCGCAGAAUAUUUUCUAAAUAUUUGUAUUUAGCGCAAGUUAAAUAAUCAGACAGAUUAC-CAAAAACUGAACAACAGCACAAA-CUGCAGA---UUCAAA-AGCG ...........(((((((....)))).....((((((((((((....))))))))(((...(((.((..-...)).))).......(((....-.))).))---).))))-.))) ( -17.30) >DroMoj_CAF1 94208 109 - 1 AUAAAAAUUGCCUGAAAAUA-UUUCUAAAUAUUUGUAUAUAGCGCAAGUUAAAUAAUCAGACAGAUUAU-GCAUACGUUAUAAAAAUAACAAA-AGCCAAAGCAGCGAAA-AA-- .......(((.(((((((((-((....))))))).(((.((((....)))).))).)))).)))....(-((....(((((....)))))...-.((....)).)))...-..-- ( -17.90) >DroAna_CAF1 84853 106 - 1 AUAAAAAUUGCCGCA-AAUAUUUUCUAAAUAUUUGUAUUUAGCGCAAGAUAAAUAAUCAGACAGAUUAUGCAUACGUUGGAC----CCAGAAA-CUAAAAA---UCCAAAAAGCA ........(((.(((-((((((.....))))))))).(((((((....((..((((((.....))))))..)).))))))).----.......-.......---........))) ( -15.40) >consensus AUAAAAAUUGCCGCAAAAUAUUUUCUAAAUAUUUGUAUUUAGCGCAAGUUAAAUAAUCAGACAGAUUAC_CAAAAACUGAACAACAGCACAAA_CUGCAAA___UCCAAA_AGCG ...............................((((((((((((....))))))))..))))...................................................... ( -8.05 = -8.38 + 0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:58 2006