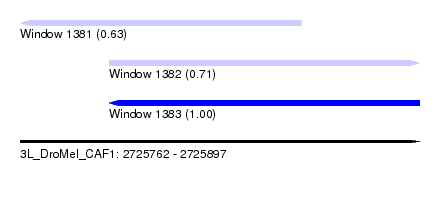
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,725,762 – 2,725,897 |
| Length | 135 |
| Max. P | 0.999907 |

| Location | 2,725,762 – 2,725,857 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 77.47 |
| Mean single sequence MFE | -15.34 |
| Consensus MFE | -8.22 |
| Energy contribution | -7.38 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.628678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2725762 95 - 23771897 GAUGCAAAUGGAAAAAGUGUGUACACAACACGAAUUAUGCAAUGGUAUAUUUG-CCCAAAAA-UUAA-----AAAAAAAAAGUUUAAAUUGAAAAGUUUACA ...(((((((......((((.......))))......(((....)))))))))-).......-....-----............((((((....)))))).. ( -12.40) >DroSim_CAF1 84106 101 - 1 GAUGCAAAUGGAAAAAGUGUGUACACAACACGAAUUAUGCAAUGGCAUAUUUG-CCCAAAAAUUCAAAAAAAAAAAAAAAAGUUUAAAUUGAAAAGUUUACA ...(((((((......((((.......))))......(((....)))))))))-).............................((((((....)))))).. ( -14.40) >DroEre_CAF1 94206 91 - 1 GAUGCAAAUGGAAAAAUUGUGUACACAACACGAAUUAUGCAAUGGCAUAUUUG-CCCAAAAA-UUCA-----A--A--AAAGUUUAAAUUGAAAAGUUUGCA ..(((((((.....((((((((.....))))(((((.......((((....))-))....))-))).-----.--.--........)))).....))))))) ( -17.60) >DroYak_CAF1 87615 93 - 1 GAUGCAAAUGGAAAAAGUGUGUACACAACACGAAUUAUGCAAUGGCAUAUUUG-CCCAAAAA-UUCA-----A--AAAAAAGUUUAAAUUGAAAAGUUUGCA ..(((((((.......((((.......))))(((((.......((((....))-))....))-))).-----.--....................))))))) ( -17.20) >DroAna_CAF1 84300 91 - 1 CUCGCAAAUCGCAAAUAAAAGUCAACAAUGCUGGU-CUUCAGCGAGAUAUUUAGUUGGAAAA-CUCG---------ACAAACUUUAAAUUAAAAAGUUUUCU ...((.....))........(((.....(((((..-...)))))........((((....))-)).)---------))(((((((.......)))))))... ( -15.10) >consensus GAUGCAAAUGGAAAAAGUGUGUACACAACACGAAUUAUGCAAUGGCAUAUUUG_CCCAAAAA_UUCA_____A__AAAAAAGUUUAAAUUGAAAAGUUUACA ........(((......(((((.....)))))((.(((((....))))).))...)))..........................((((((....)))))).. ( -8.22 = -7.38 + -0.84)

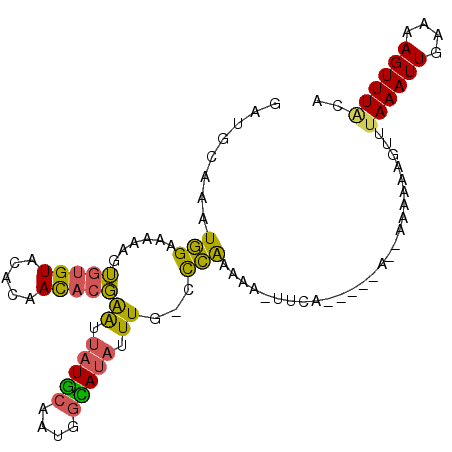
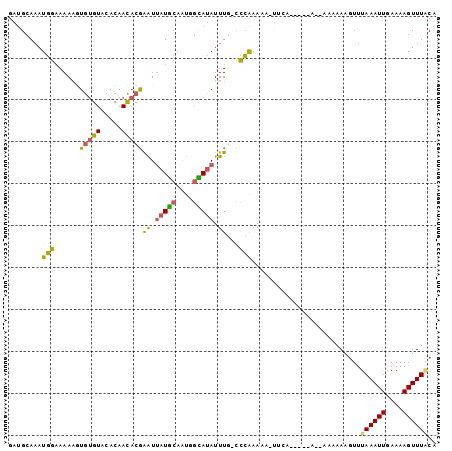
| Location | 2,725,792 – 2,725,897 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -15.53 |
| Consensus MFE | -14.30 |
| Energy contribution | -14.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.714082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2725792 105 + 23771897 ----UUAA-UUUUUGGGCAAAUAUACCAUUGCAUAAUUCGUGUUGUGUACACACUUUUUCCAUUUGCAUCGUCUCAUUUUAACUUUUCCCUUUGAUUCGCCCCGCCUUUU ----....-.....((((...........((((((((....)))))))).....................(..(((................)))..)))))........ ( -13.69) >DroSec_CAF1 85923 109 + 1 UUUUUUGA-UUUUUGGGCAAAUAUGCCAUUGCAUAAUUCGUGUUGUGUACACACUUUUUCCAUUUGCAUCGUCUCAUUUUAACUUUUCCCUUUGAUUCGCCCCGCCUUUU ........-.....((((...(((((....)))))....((((.....))))..............................................))))........ ( -15.60) >DroSim_CAF1 84137 110 + 1 UUUUUUGAAUUUUUGGGCAAAUAUGCCAUUGCAUAAUUCGUGUUGUGUACACACUUUUUCCAUUUGCAUCGUCUCAUUUUAACUUUUCCCUUUGAUUCGCCCCGCCUUUU ......(((((...(((.((((((((....)))))....((((.....))))...............................))).)))...)))))............ ( -16.40) >DroEre_CAF1 94232 105 + 1 ----UGAA-UUUUUGGGCAAAUAUGCCAUUGCAUAAUUCGUGUUGUGUACACAAUUUUUCCAUUUGCAUCGUCUCAUUUUAACUUUCUCCUUUGAUUCGCCCCGCCUUUU ----.(((-((...((((((((((((....)))))....((((.....))))..........)))))...((.........)).....))...)))))............ ( -15.90) >DroYak_CAF1 87643 105 + 1 ----UGAA-UUUUUGGGCAAAUAUGCCAUUGCAUAAUUCGUGUUGUGUACACACUUUUUCCAUUUGCAUCGUCUCAUUUUAACUUUUUCCUUUGACUCGCCCCGCCAUUU ----....-.....((((...(((((....)))))....((((.....))))..................(((....................)))..))))........ ( -16.05) >consensus ____UUAA_UUUUUGGGCAAAUAUGCCAUUGCAUAAUUCGUGUUGUGUACACACUUUUUCCAUUUGCAUCGUCUCAUUUUAACUUUUCCCUUUGAUUCGCCCCGCCUUUU ..............((((...(((((....)))))....((((.....))))..............................................))))........ (-14.30 = -14.50 + 0.20)

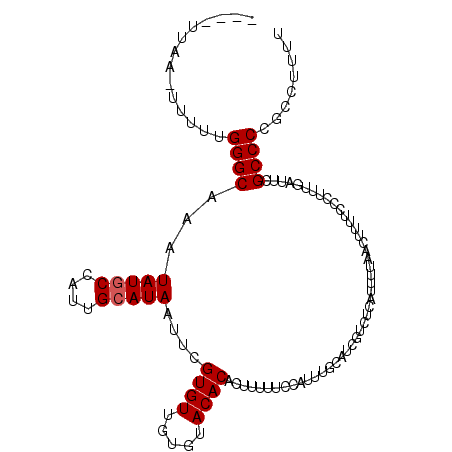
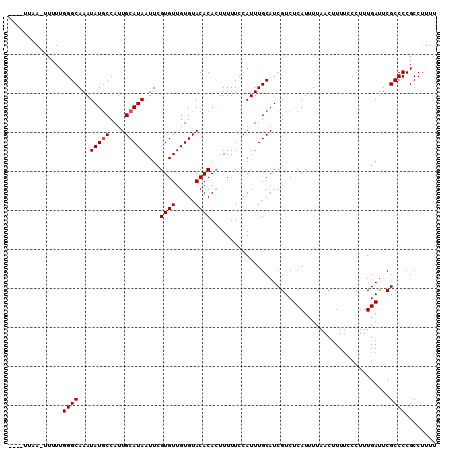
| Location | 2,725,792 – 2,725,897 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -20.62 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.30 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.20 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
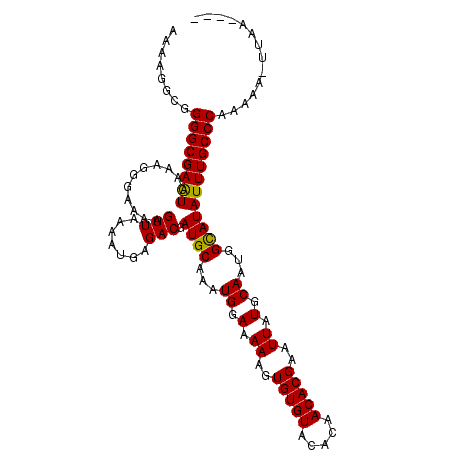
>3L_DroMel_CAF1 2725792 105 - 23771897 AAAAGGCGGGGCGAAUCAAAGGGAAAAGUUAAAAUGAGACGAUGCAAAUGGAAAAAGUGUGUACACAACACGAAUUAUGCAAUGGUAUAUUUGCCCAAAAA-UUAA---- ........((((((((...........(((.......))).((((...((.(.((..(((((.....)))))..)).).))...)))))))))))).....-....---- ( -18.80) >DroSec_CAF1 85923 109 - 1 AAAAGGCGGGGCGAAUCAAAGGGAAAAGUUAAAAUGAGACGAUGCAAAUGGAAAAAGUGUGUACACAACACGAAUUAUGCAAUGGCAUAUUUGCCCAAAAA-UCAAAAAA ........((((((((...........(((.......))).((((...((.(.((..(((((.....)))))..)).).))...)))))))))))).....-........ ( -20.80) >DroSim_CAF1 84137 110 - 1 AAAAGGCGGGGCGAAUCAAAGGGAAAAGUUAAAAUGAGACGAUGCAAAUGGAAAAAGUGUGUACACAACACGAAUUAUGCAAUGGCAUAUUUGCCCAAAAAUUCAAAAAA ........((((((((...........(((.......))).((((...((.(.((..(((((.....)))))..)).).))...)))))))))))).............. ( -20.80) >DroEre_CAF1 94232 105 - 1 AAAAGGCGGGGCGAAUCAAAGGAGAAAGUUAAAAUGAGACGAUGCAAAUGGAAAAAUUGUGUACACAACACGAAUUAUGCAAUGGCAUAUUUGCCCAAAAA-UUCA---- ........((((((((...........(((.......))).((((...((.(.((.((((((.....)))))).)).).))...)))))))))))).....-....---- ( -21.90) >DroYak_CAF1 87643 105 - 1 AAAUGGCGGGGCGAGUCAAAGGAAAAAGUUAAAAUGAGACGAUGCAAAUGGAAAAAGUGUGUACACAACACGAAUUAUGCAAUGGCAUAUUUGCCCAAAAA-UUCA---- ........((((((((...........(((.......))).((((...((.(.((..(((((.....)))))..)).).))...)))))))))))).....-....---- ( -20.80) >consensus AAAAGGCGGGGCGAAUCAAAGGGAAAAGUUAAAAUGAGACGAUGCAAAUGGAAAAAGUGUGUACACAACACGAAUUAUGCAAUGGCAUAUUUGCCCAAAAA_UUAA____ ........((((((((...........(((.......))).((((...((.(.((..(((((.....)))))..)).).))...)))))))))))).............. (-20.62 = -20.30 + -0.32)

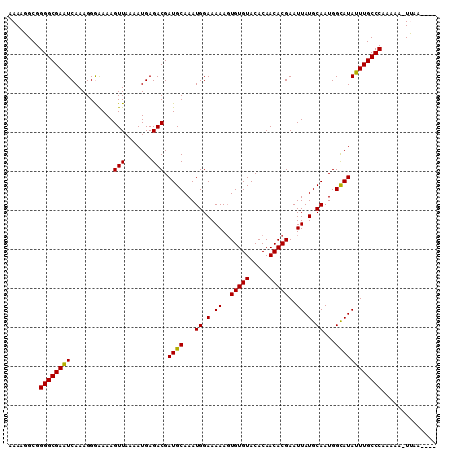
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:55 2006