
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,718,882 – 2,718,984 |
| Length | 102 |
| Max. P | 0.510849 |

| Location | 2,718,882 – 2,718,984 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.25 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -7.77 |
| Energy contribution | -9.80 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.42 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2718882 102 + 23771897 GGUUCAGUGCUACCUGAACCAAACAAUUGCUCGACAGCAAAAGUCAAUUGGGCG---ACACACGCAACAUCACACACACACAC---UCA-GCACAGCACAGCACACCAC (((...(((((..((((.((((((..(((((....)))))..))...))))(((---.....)))..................---)))-)...))))).....))).. ( -22.50) >DroVir_CAF1 93693 86 + 1 CCUUUUGUGCUACCUGA-CCAAACAAUUGCACCACAACAAAAGUCAAUUGGACGGACACACACACAAAUUCGAACACACG-----------CAUAG-----------GC ...((((((.....((.-((...((((((.((..........))))))))...)).))....))))))............-----------.....-----------.. ( -11.00) >DroSec_CAF1 79250 95 + 1 GGUUCAGUGCUACCUGAACCAAACAAUUGCUCGACAGCAAAAGUCAAUUGGGCG---ACACACGCAACAGCACACACGCU-----------CACAGCGCAGCAAGCCAC (((((((......)))))))......(((((((((.......)))...((((((---......((....)).....))))-----------))....).)))))..... ( -25.50) >DroSim_CAF1 78195 93 + 1 GGUUCAGUGCUACCUGAACCAAACAAUUGCUCGACAGCAAAAGUCAAUUGGGCG---ACACACGCAACAGCACACACA-------------CACAGCGCAGCAAGCCAC (((((((......)))))))...((((((((..........)).))))))(((.---.....(((.............-------------....)))......))).. ( -21.63) >DroEre_CAF1 86598 88 + 1 GGUUCACUGCCACCUGAACCAAACAAUUGCUCGACAGCAAAAGUCAAUUGGGCG---ACACACGCAACAUCACACACACACCCCAAACACC------------------ ((((((........))))))......((((((((..((....))...)))))))---).................................------------------ ( -14.60) >DroAna_CAF1 79220 84 + 1 -AUUCUCGACUACCUGAACCAAACAAUUGCUCGACACCAAAAGUCAAUUGGGCAGGCAUACACGCA-CAGCACAGGC--CC----------CACAGAG----------- -...(((.....((((..........(((((((((.......)))....))))))((......)).-.....)))).--..----------....)))----------- ( -14.60) >consensus GGUUCAGUGCUACCUGAACCAAACAAUUGCUCGACAGCAAAAGUCAAUUGGGCG___ACACACGCAACAGCACACACACA___________CACAGCG_________AC (((((((......)))))))...((((((((..........)).)))))).(((........)))............................................ ( -7.77 = -9.80 + 2.03)

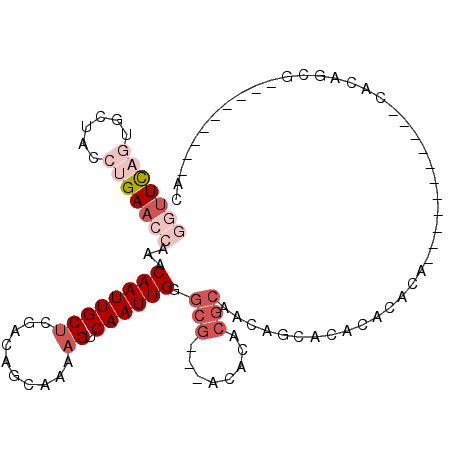
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:53 2006