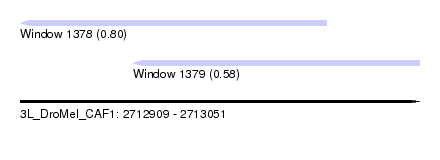
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,712,909 – 2,713,051 |
| Length | 142 |
| Max. P | 0.802868 |

| Location | 2,712,909 – 2,713,018 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -36.47 |
| Consensus MFE | -32.22 |
| Energy contribution | -32.58 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
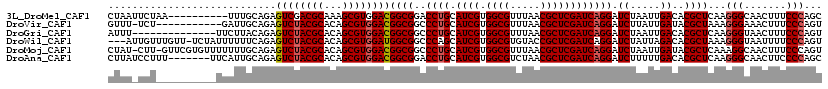
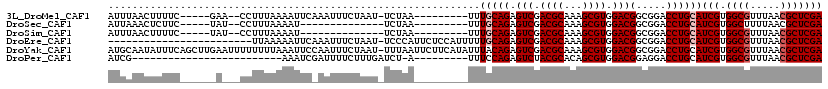
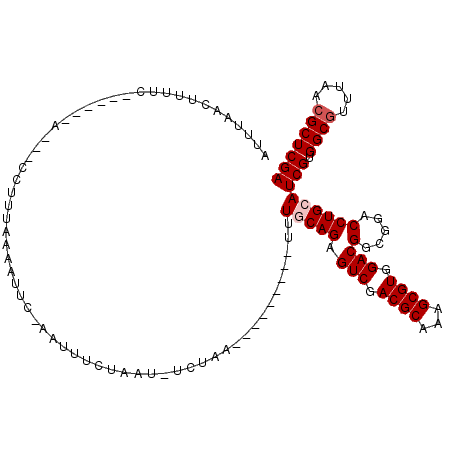
>3L_DroMel_CAF1 2712909 109 - 23771897 CUAAUUCUAA----------UUUGCAGAGUCGACGCAAAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGCAACUUUCCCAGC ..........----------(((((...(((.((((...)))).))).)))))((((.((((.((((.....)))))))))))).............(((.((((....))))...))) ( -34.40) >DroVir_CAF1 88057 107 - 1 GUUU-UCU-----------GAUUGCAGAGUCUACGCACAGCGUGGACGGCGGCCCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUUAUUGAUACGCUAAAGGGAAACUUUCCCAGU ....-(((-----------(....))))((((((((...))))))))((((..((((.((((.((((.....))))))))))))(((.....))).))))...((((.....))))... ( -41.70) >DroGri_CAF1 79083 105 - 1 AUUU--------------UUCUUACAGAGUCUACGCACAGCGUGGACGGCGGCCCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGUAACUUUCCCAGU ....--------------..........((((((((...))))))))((((..((((.((((.((((.....)))))))))))).((.....))..))))...(((.......)))... ( -35.40) >DroWil_CAF1 89674 115 - 1 ---AUUGUUUGUU-UCUAUUUUUUCAGAGUCUACGCACAGCGUGGAUGGCGGCCCAGCAUCGUGGCGUGUACCGCUCGAUCAGGAUCUAUUAGACACGCUAAAGGGUAAUUUUCCCAGU ---.......((.-((((....(((.((((((((((...))))))))(((((..((((......)).))..)))))...)).))).....)))).))......(((.......)))... ( -29.80) >DroMoj_CAF1 79619 117 - 1 CUAU-CUU-GUUCGUGUUUUUUUGCAGAGUCUACGCACAGCGUGGACGGCGGCCCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGAUACGCUCAAAGGCAACUUUCCCAGU ....-..(-(..((((((..((.((...((((((((...)))))))).))(..((((.((((.((((.....))))))))))))..).))..))))))..))(((....)))....... ( -39.70) >DroAna_CAF1 73019 112 - 1 CUUAUCCUUU-------UUCAUUGCAGAGUCUACGCACAGCGUGGACGGCGGACCUGCAUCGUGGCGUCUAACGCUCGAUCAGGAUCUUUUUGACACGCUCAAGGGCAACUUCCCCAGC ..........-------......((...((((((((...))))))))((.(((((((.((((.((((.....)))))))))))).............(((....)))....))))).)) ( -37.80) >consensus CUUA_UCU__________UUUUUGCAGAGUCUACGCACAGCGUGGACGGCGGCCCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGCAACUUUCCCAGU ............................((((((((...))))))))((((..((((.((((.((((.....)))))))))))).((.....))..))))...(((.......)))... (-32.22 = -32.58 + 0.36)
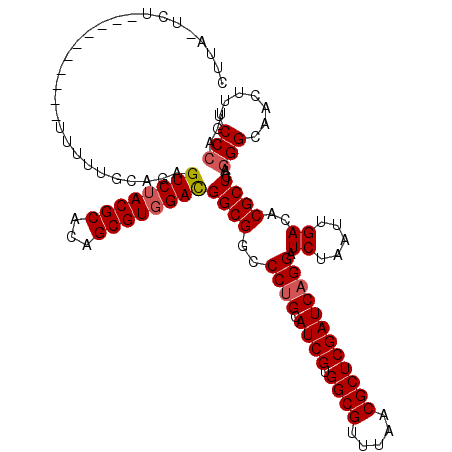
| Location | 2,712,949 – 2,713,051 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.22 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.31 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2712949 102 - 23771897 AUUUAACUUUUC-----GAA--CCUUUAAAAUUCAAAUUUCUAAU-UCUAA---------UUUGCAGAGUCGACGCAAAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGA ............-----...--.......................-.....---------..(((((.(((.((((...)))).)))(.....))))))(((.((((.....))))))) ( -21.00) >DroSec_CAF1 73638 89 - 1 AUUAAACUCUUC-----UAU--CCUUUAAAAU--------------UCUAA---------UUUGCAGAGUCGACGCAAAGCGUGGACGGCGGACCUGCAUCGUGGCUUUUAACGCUCGA ............-----...--...((((((.--------------.(((.---------..(((((.(((.((((...)))).)))(.....))))))...))).))))))....... ( -17.80) >DroSim_CAF1 71125 89 - 1 AUUUAACUUUUC-----UAU--CCUUUAAAAU--------------UCUAA---------UUUGCAGAGUCGACGCAAAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGA ............-----...--..........--------------.....---------..(((((.(((.((((...)))).)))(.....))))))(((.((((.....))))))) ( -21.00) >DroEre_CAF1 80830 94 - 1 ------------------------UUAAAAAUUCAAAUUUCUAAU-UCCCAUUCUCCAUUUUUGCAGAGUCGACGCAAAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGA ------------------------....................(-((((..(((.((....)).)))(((.((((...)))).))))).)))......(((.((((.....))))))) ( -21.10) >DroYak_CAF1 74430 118 - 1 AUGCAAUAUUUCAGCUUGAAUUUUUUUUAAAUUCCAAUUUCUAAU-UUUAAUUCUUCAUAUUUACAGAGUCGACGCAAAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGA .((((....(((.(((.((((((.....))))))...........-......................(((.((((...)))).)))))))))..))))(((.((((.....))))))) ( -24.30) >DroPer_CAF1 97725 84 - 1 AUCG-------------------------AAAUCGAUUUUCUUUGAUCU-A---------UUUCCAGAGUCUACGCACAGCGUGGACGGAGGACCUGCAUCGUGGCGUUUAACGCUCGA ...(-------------------------((((.((((......)))).-)---------))))....((((((((...))))))))((....))....(((.((((.....))))))) ( -24.20) >consensus AUUUAACUUUUC______A___CCUUUAAAAUUC_AAUUUCUAAU_UCUAA_________UUUGCAGAGUCGACGCAAAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGA ..............................................................(((((.(((.((((...)))).)))(.....))))))(((.((((.....))))))) (-17.60 = -18.10 + 0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:52 2006