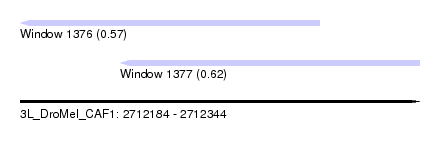
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,712,184 – 2,712,344 |
| Length | 160 |
| Max. P | 0.618250 |

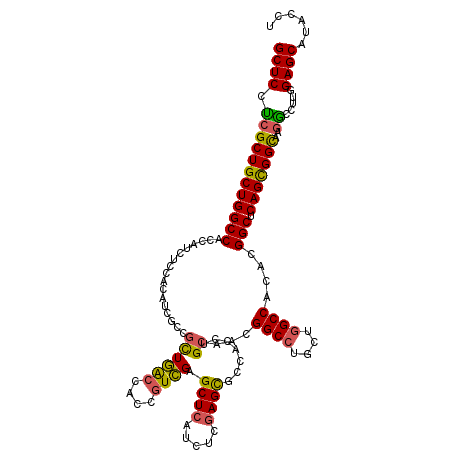
| Location | 2,712,184 – 2,712,304 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.56 |
| Mean single sequence MFE | -44.83 |
| Consensus MFE | -30.99 |
| Energy contribution | -31.13 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.571492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2712184 120 - 23771897 CACCGUCGAGCUCAUCUCGAGCGCCGCCUGCAGUGGCCUCCUGGCCACCAGGCUCAGCGGCAAGGAUUUGGAGCACUCCUUUCAGGACAUAAUAUUGCUGCACGAGGCCUACGACGACAU ...(((((.((((.....))))((((((((..((((((....)))))))))))...(((((((.((...(((....)))..))...........)))))))....)))...))))).... ( -52.00) >DroVir_CAF1 87314 120 - 1 CACCGUCGAGCUCAUCUCGAGCGACAGCUGCAACGGCCUGCUGGCCACACGGCUCAGCGGCAAGGCCUUGGAGCAUAAUUUUCAGAACAUAAUACUGCUGCAAGAGGCCUACGACGAUAU ...(((((.((((.....)))).............(((.(((((((....))).))))))).((((((((.((((..(((..........)))..)))).))..)))))).))))).... ( -44.60) >DroGri_CAF1 78364 120 - 1 CACCGUCGAGCUCAUCUCGAGUGCCACUUGCAACGGCCUGCUGGCCACACGGCUCAGCGGUAAGGCCUUGGAGCAUACUUUUCAGGACAUCAUACUGCUGCAAGAGGCAUUCGACACUAU ....((((((.....)))(((((((.((((((.(((.(((((((((....))).))))))......(((((((......)))))))........))).)))))).))))))))))..... ( -48.00) >DroWil_CAF1 88919 118 - 1 --CCGUUGAGCUAAUUUCGAGUGCCGCCUGCAACGGCCUGCUGGCCACCAGGCUCAGCGGCAAGGCCCUAGAGCAUACAUUUCAGGACAUCAUAUUACUGCACGAGGCCUUCGACGAUAU --.((((((............(((((((......)))..(((((((....))).))))))))(((((...(.(((.......................))).)..))))))))))).... ( -37.70) >DroMoj_CAF1 78900 120 - 1 CACCGUCGAGCUCAUCUCGAGCGGCACCUGCAACGGCCUGCUGGCCACACGGCUCAGCGGCAAAGCCUUGGAGCAUACCUUUCAGGAUAUAAUAUUCCUGCACGACGCCUACGACGAUAU ...(((((.((((.....))))(((..((.(((.((((((((((((....))).))))))....)))))).)).........(((((........)))))......)))..))))).... ( -46.40) >DroAna_CAF1 72267 120 - 1 CACCGUCGAGCUCAUUUCGAGCGCCGCCUGCGGGGGCCUCCUCGCCACCCGGCUCAGUGGCAAGACCUUGGAGCACACCUUUCAAGAUAUAAUACUGCUGCACGAGACCUACGACGACAU ...(((((((....(((((.(((((((..((((((((......))).))).))...))))).....(((((((......))))))).............)).))))).)).))))).... ( -40.30) >consensus CACCGUCGAGCUCAUCUCGAGCGCCACCUGCAACGGCCUGCUGGCCACACGGCUCAGCGGCAAGGCCUUGGAGCAUACCUUUCAGGACAUAAUACUGCUGCACGAGGCCUACGACGAUAU ...(((((.((((.....))))......((((...(((.(((((((....))).))))))).....(((((((......)))))))............)))).........))))).... (-30.99 = -31.13 + 0.14)

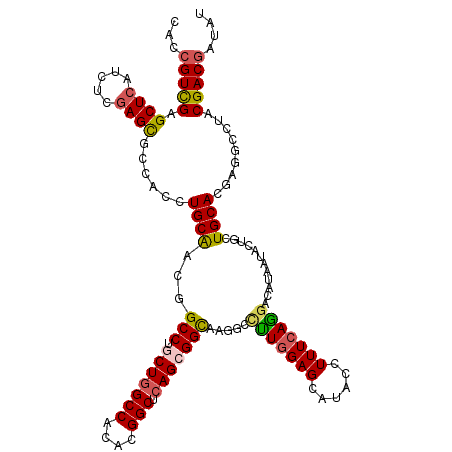
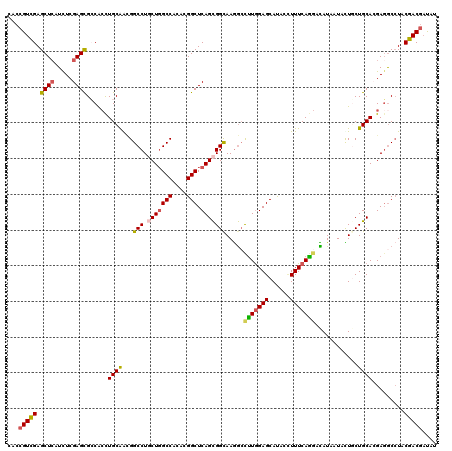
| Location | 2,712,224 – 2,712,344 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -46.42 |
| Consensus MFE | -35.21 |
| Energy contribution | -34.72 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.618250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2712224 120 - 23771897 GCUCCUCGCUGCUGGCCACCAUCUCCACGUCGCCACUGACCACCGUCGAGCUCAUCUCGAGCGCCGCCUGCAGUGGCCUCCUGGCCACCAGGCUCAGCGGCAAGGAUUUGGAGCACUCCU ..((((.(((((((((............((((....)))).........((((.....)))))))(((((..((((((....)))))))))))..)))))).))))...(((....))). ( -50.30) >DroVir_CAF1 87354 120 - 1 GCUCUUCGCUGCUGGCCACCUUCUCCACAUCGCCGUUGACCACCGUCGAGCUCAUCUCGAGCGACAGCUGCAACGGCCUGCUGGCCACACGGCUCAGCGGCAAGGCCUUGGAGCAUAAUU (((((..(((....(((..............(((((((.(..(.(((..((((.....))))))).)..))))))))..(((((((....))).)))))))..)))...)))))...... ( -47.90) >DroGri_CAF1 78404 120 - 1 GCUCAUCGCUGCUGGCCACCAUCUCCACAUCGCCGCUGACCACCGUCGAGCUCAUCUCGAGUGCCACUUGCAACGGCCUGCUGGCCACACGGCUCAGCGGUAAGGCCUUGGAGCAUACUU .......(((.(.((((...((......)).((((((((((....(((((.....))))).(((.....)))..((((....))))....)).))))))))..))))..).)))...... ( -42.00) >DroWil_CAF1 88959 117 - 1 GCUCCCCGCUGCUGGCCACCAUAUCGAAAUCACCGCUAA---CCGUUGAGCUAAUUUCGAGUGCCGCCUGCAACGGCCUGCUGGCCACCAGGCUCAGCGGCAAGGCCCUAGAGCAUACAU ((((((.(((((((((.((....(((((((....(((..---......)))..))))))))))))(((((....((((....))))..)))))..))))))..)).....))))...... ( -42.40) >DroMoj_CAF1 78940 120 - 1 GCUCAUCGCUGCUGGCCACCAUCUCGUCAUCACCGCUGGCCACCGUCGAGCUCAUCUCGAGCGGCACCUGCAACGGCCUGCUGGCCACACGGCUCAGCGGCAAAGCCUUGGAGCAUACCU ((((...(((((((((((.(..............).))))))...(((((.....)))))))))).....(((.((((((((((((....))).))))))....))))))))))...... ( -47.44) >DroAna_CAF1 72307 120 - 1 GCUCCUCGCUGCUGGCCACCAUCUCCGCCUCGCCGCUGACCACCGUCGAGCUCAUUUCGAGCGCCGCCUGCGGGGGCCUCCUCGCCACCCGGCUCAGUGGCAAGACCUUGGAGCACACCU (((((......(((((..........)))..((((((((..........((((.....))))((((...((((((....))))))....)))))))))))).)).....)))))...... ( -48.50) >consensus GCUCCUCGCUGCUGGCCACCAUCUCCACAUCGCCGCUGACCACCGUCGAGCUCAUCUCGAGCGCCACCUGCAACGGCCUGCUGGCCACACGGCUCAGCGGCAAGGCCUUGGAGCAUACCU ((((.((((((((((((.................((((((....)))).((((.....)))).......))...((((....))))....))).)))))))..)).....))))...... (-35.21 = -34.72 + -0.49)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:50 2006