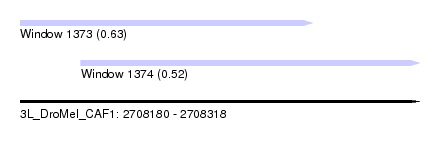
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,708,180 – 2,708,318 |
| Length | 138 |
| Max. P | 0.629126 |

| Location | 2,708,180 – 2,708,281 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -29.95 |
| Consensus MFE | -19.85 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2708180 101 + 23771897 AAUCUG---CUUUUCGGAACCCUCUACUUACAUGGUAUGAUGCCCUGCUCGACGAGCUGCGAGUGUGGACGUCGCACCAUGAGCUUCACUUUGAGCGCUGCAAA ....((---(....................((((((.(((((((.((((((........)))))).)).))))).))))))((((((.....))).)))))).. ( -30.00) >DroGri_CAF1 73676 93 + 1 ACUUGUUGC-----------GAUGCACUUACAUGGUAUAAUGCCCUGCUCGACGAGUUGGGAGUGCGGCCGUCGCACCAUCAGUUUCACUUUGAGCGCUGCAAA ...((.(((-----------((((((((..(((......)))(((.((((...)))).))))))))....)))))).)).(((((((.....))).)))).... ( -25.40) >DroSec_CAF1 68964 101 + 1 AAUUUG---CUUUUCGGACCCCUCUACUUACAUGGUAUGAUGCCCUGCUCGACGAGCUGCGAGUGUGGACGUCGCACCAUAAGCUUCACUUUGAGCGCUGCAAA ..((((---(.....................(((((.(((((((.((((((........)))))).)).))))).))))).((((((.....))).)))))))) ( -27.90) >DroWil_CAF1 82626 94 + 1 GAUUUC---UUUUU------CUUUUACUUACAUGGUAUGAUGCCCUGCUCGACAAGUUGUGAGUGCGGCCUCCGCACCAUCAGCUUCACUUUGAGCGCUGCAA- ......---....(------(...((((.....)))).))(((..(((((((.((((((((.((((((...)))))))).))))))....)))))))..))).- ( -29.00) >DroMoj_CAF1 74419 93 + 1 ACGCAGCGU-----------GGUGCACUUACAUGGUAUAAUGCCCUGUUCGACAAGUUGCGAGUGCGGCCGUCGCACCAUCAGCUUCACUUUGAGCGCUGCAAG ..(((((((-----------(((((....(((.(((.....))).)))..(((..(((((....))))).))))))))))..((((......)))))))))... ( -36.80) >DroAna_CAF1 68683 89 + 1 AUUCUG---------G-----CGAUACUUACAUGGUAUGAUGCCCUGCUCCACGAGCUGCGAGUGUGGCCUCCGCACCAUGAGCUUCACUUUGAGCGCUGGAA- .(((..---------(-----(........((((((..((.(((.(((((((.....)).))))).))).))...)))))).((((......))))))..)))- ( -30.60) >consensus AAUCUG______________CCUCUACUUACAUGGUAUGAUGCCCUGCUCGACGAGCUGCGAGUGCGGCCGUCGCACCAUCAGCUUCACUUUGAGCGCUGCAAA ........................((((.....))))...(((..(((((((.(((((....((((((...))))))....)))))....)))))))..))).. (-19.85 = -19.30 + -0.55)

| Location | 2,708,201 – 2,708,318 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.67 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -18.06 |
| Energy contribution | -19.17 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2708201 117 + 23771897 UACUUACAUGGUAUGAUGCCCUGCUCGACGAGCUGCGAGUGUGGACGUCGCACCAUGAGCUUCACUUUGAGCGCUGCAAA-AUAUG--GAGAACCAAAGAAGCAAGUUAGUUAAGCAACA .((((.((((((.(((((((.((((((........)))))).)).))))).)))))).(((((..((((.(..((.((..-...))--.))..))))))))))))))............. ( -35.60) >DroPse_CAF1 89672 107 + 1 UACUUACAUGGUAUGAUGCCCUGUUCGACCAACUGCGAGUGGGGUCGUCGCACCAUGAGCUUCACUUUGAGCGCUGCAAA-AAAUA--UA----CAAAGAUACAAUUUAU---AGCA--- ......((((((.((((((((..((((........))))..))).))))).)))))).((((......))))((((.(((-...((--(.----.....)))...))).)---))).--- ( -33.80) >DroGri_CAF1 73689 110 + 1 CACUUACAUGGUAUAAUGCCCUGCUCGACGAGUUGGGAGUGCGGCCGUCGCACCAUCAGUUUCACUUUGAGCGCUGCAAAGAAAUC--GAUAAAGAAAUGAUCAAAUUAUU-----A--- ........((((....(((..(((((((.((..(((..(((((.....)))))..)))..))....)))))))..)))......((--......))....)))).......-----.--- ( -23.00) >DroMoj_CAF1 74432 112 + 1 CACUUACAUGGUAUAAUGCCCUGUUCGACAAGUUGCGAGUGCGGCCGUCGCACCAUCAGCUUCACUUUGAGCGCUGCAAGUAGAGAGAAAUCAUAAAAUAAUACAAUCAUU-----A--- ..........((((..(((..(((((((.((((((...(((((.....)))))...))))))....)))))))..)))......((....))........)))).......-----.--- ( -25.10) >DroAna_CAF1 68693 95 + 1 UACUUACAUGGUAUGAUGCCCUGCUCCACGAGCUGCGAGUGUGGCCUCCGCACCAUGAGCUUCACUUUGAGCGCUGGAA--AGAUA--CAGUU---------CAGGUU------------ .((((.((((((..((.(((.(((((((.....)).))))).))).))...)))))).((((......))))((((...--.....--)))).---------.)))).------------ ( -26.10) >DroPer_CAF1 90511 107 + 1 UACUUACAUGGUAUGAUGCCCUGUUCGACCAACUGCGAGUGGGGUCGUCGCACCAUGAGCUUCACUUUUAGCGCUGCAAA-AAAUA--UA----CAAAGAUACAAUUUAU---AGCA--- ......((((((.((((((((..((((........))))..))).))))).)))))).(((........)))((((.(((-...((--(.----.....)))...))).)---))).--- ( -33.10) >consensus UACUUACAUGGUAUGAUGCCCUGCUCGACGAGCUGCGAGUGCGGCCGUCGCACCAUGAGCUUCACUUUGAGCGCUGCAAA_AAAUA__GA__A_CAAAGAAACAAAUUAU______A___ ......((((((.(((((((.((((((........)))))).)).))))).)))))).((((......))))................................................ (-18.06 = -19.17 + 1.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:47 2006