| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,696,644 – 2,696,778 |
| Length | 134 |
| Max. P | 0.821840 |

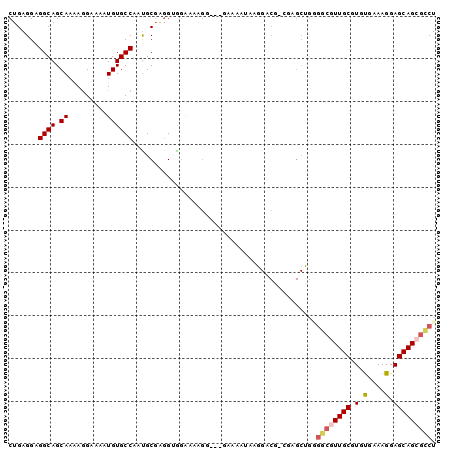
| Location | 2,696,644 – 2,696,740 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -23.18 |
| Consensus MFE | -15.44 |
| Energy contribution | -16.17 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2696644 96 + 23771897 CUGAGGAGGCAGCAAAAGGAAAAUGUGCCAACGCGAGGUGGAAAAGG---GAAAAUAAGGACG-CGAGCUGAGGCGUUGCGUGUGAAAGGAGCAGCGUCU .........((((....((........))..((((............---...........))-)).))))(((((((((.(.(....).)))))))))) ( -23.50) >DroSec_CAF1 57503 96 + 1 CUGAGGAGGCAGCAAAAGGAAAAUGUGCCAAUGCGAGGUGGAAAAGG---GAAAAUAAGGACG-CGAGCUGGGGCGUUGCGUGCGAAAGGAGCAGCGCCU .........((((....((........))..((((............---...........))-)).))))(((((((((.(.(....).)))))))))) ( -27.40) >DroSim_CAF1 56390 96 + 1 CUGAGGAGGCAGCAAAAGGAAAAUGUGCCAAUGCGAGGAGGAAAAGG---GAAAAUAAGGACG-CGAGCUGGGGCGUUGCGUGUGAAAGGAGCAGCGCCU .........((((....((........))..((((............---...........))-)).))))(((((((((.(.(....).)))))))))) ( -24.40) >DroEre_CAF1 62408 94 + 1 CUGAGGAGGCAGCAAAAGGAAAAUGUGCCAAUGCGAGGUGGAAAAGG---GAAAA--AGGACG-AGAGCUGGGGCGUUGCGUGUGAAAGGAGCAGCGCCU .........((((..........(.((((.......)))).)...(.---.....--....).-...))))(((((((((.(.(....).)))))))))) ( -21.90) >DroAna_CAF1 56084 100 + 1 CUGAGGAGGCAGCAAAAAGAAAAUGUGCCAAUGCGAUGAACAAAGGGAGGGAGGAAACCGUCAAGGGGCGGCGGCAUUGCGUGUGAAAGGAGCAGCGCCU ......((((.((.........(..(((.(((((..((..(.....((.((......)).))...)..))...))))))))..)..........)))))) ( -22.81) >DroPer_CAF1 74486 73 + 1 CUGAGGUGGCAGCAAGAGGAAAAUGUGCCAAUGCCGGGCAGGGCAUG---G------------------------AUUGCGUGUGAAAGGAGCAGCGGCU ....(((.((.((.........((((.(((.((((......))))))---)------------------------...)))).........)).)).))) ( -19.07) >consensus CUGAGGAGGCAGCAAAAGGAAAAUGUGCCAAUGCGAGGUGGAAAAGG___GAAAAUAAGGACG_CGAGCUGGGGCGUUGCGUGUGAAAGGAGCAGCGCCU .......((((.((.........))))))...........................................((((((((...........)))))))). (-15.44 = -16.17 + 0.72)


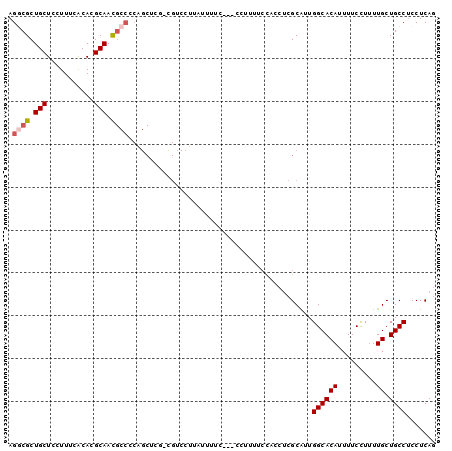
| Location | 2,696,644 – 2,696,740 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.70 |
| Mean single sequence MFE | -15.71 |
| Consensus MFE | -11.10 |
| Energy contribution | -11.55 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.71 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2696644 96 - 23771897 AGACGCUGCUCCUUUCACACGCAACGCCUCAGCUCG-CGUCCUUAUUUUC---CCUUUUCCACCUCGCGUUGGCACAUUUUCCUUUUGCUGCCUCCUCAG .((((((((...........)))((((........)-)))..........---.............)))))((((((.........)).))))....... ( -15.80) >DroSec_CAF1 57503 96 - 1 AGGCGCUGCUCCUUUCGCACGCAACGCCCCAGCUCG-CGUCCUUAUUUUC---CCUUUUCCACCUCGCAUUGGCACAUUUUCCUUUUGCUGCCUCCUCAG (((((((((.......))).)).((((........)-)))..........---.............(((..((........))...))).))))...... ( -16.60) >DroSim_CAF1 56390 96 - 1 AGGCGCUGCUCCUUUCACACGCAACGCCCCAGCUCG-CGUCCUUAUUUUC---CCUUUUCCUCCUCGCAUUGGCACAUUUUCCUUUUGCUGCCUCCUCAG .((((.(((...........))).)))).......(-((...........---............)))...((((((.........)).))))....... ( -16.40) >DroEre_CAF1 62408 94 - 1 AGGCGCUGCUCCUUUCACACGCAACGCCCCAGCUCU-CGUCCU--UUUUC---CCUUUUCCACCUCGCAUUGGCACAUUUUCCUUUUGCUGCCUCCUCAG .((((.(((...........))).))))........-......--.....---..................((((((.........)).))))....... ( -14.50) >DroAna_CAF1 56084 100 - 1 AGGCGCUGCUCCUUUCACACGCAAUGCCGCCGCCCCUUGACGGUUUCCUCCCUCCCUUUGUUCAUCGCAUUGGCACAUUUUCUUUUUGCUGCCUCCUCAG .((((.(((...........))).))))((((........))))...........................((((((.........)).))))....... ( -17.40) >DroPer_CAF1 74486 73 - 1 AGCCGCUGCUCCUUUCACACGCAAU------------------------C---CAUGCCCUGCCCGGCAUUGGCACAUUUUCCUCUUGCUGCCACCUCAG .((((..((...........(((..------------------------.---..)))...)).))))..(((((((.........)).)))))...... ( -13.54) >consensus AGGCGCUGCUCCUUUCACACGCAACGCCCCAGCUCG_CGUCCUUAUUUUC___CCUUUUCCACCUCGCAUUGGCACAUUUUCCUUUUGCUGCCUCCUCAG .((((.(((...........))).))))...........................................((((((.........)).))))....... (-11.10 = -11.55 + 0.45)


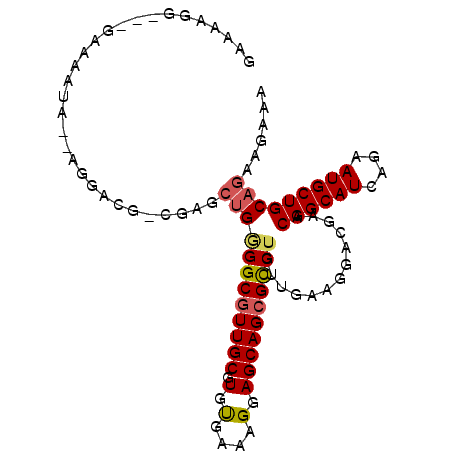
| Location | 2,696,684 – 2,696,778 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 88.26 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -20.67 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821840 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2696684 94 + 23771897 GAAAAGG---GAAAAUA--AGGACG-CGAGCUGAGGCGUUGCGUGUGAAAGGAGCAGCGUCUGUUGAAGGACGAACGGCAUCAGAAUGCUGCAGAAGAAA .......---.......--.(.(((-(((((....)).)))))).).......((((((((((.((..(......)..)).))).)))))))........ ( -26.60) >DroSec_CAF1 57543 94 + 1 GAAAAGG---GAAAAUA--AGGACG-CGAGCUGGGGCGUUGCGUGCGAAAGGAGCAGCGCCUGUUGAAGGUCGAACGGCAUCAGAAUGCUGCAGGAGAAA .......---.......--......-(((.((.(((((((((.(.(....).)))))))))).....)).)))..((((((....))))))......... ( -29.20) >DroSim_CAF1 56430 94 + 1 GAAAAGG---GAAAAUA--AGGACG-CGAGCUGGGGCGUUGCGUGUGAAAGGAGCAGCGCCUGUUGAAGGACGAACGGCAUCAGAAUGCUGCAGAAGAAA .......---.......--......-....((((((((((((.(.(....).)))))))))).............((((((....)))))))))...... ( -24.30) >DroEre_CAF1 62448 92 + 1 GAAAAGG---GAAAA----AGGACG-AGAGCUGGGGCGUUGCGUGUGAAAGGAGCAGCGCCUGUUGAAGGACGAACGGCAUCAGAAUGCUGCAGAAGAAA .......---.....----......-....((((((((((((.(.(....).)))))))))).............((((((....)))))))))...... ( -24.30) >DroYak_CAF1 58101 96 + 1 GAAAAGG---GAAAAAAAGAGGACG-AGAGCUGGGGCGUUGCGUGUGAAAGGAGCAGCGCCUGUUGAAGGACGAACGGCAUUAGAAUGCUGCAGAAGAAA .......---...............-....((((((((((((.(.(....).)))))))))).............((((((....)))))))))...... ( -24.40) >DroAna_CAF1 56124 98 + 1 CAAAGGGAGGGAGGAAA--CCGUCAAGGGGCGGCGGCAUUGCGUGUGAAAGGAGCAGCGCCUAUUGAAGGACGAACGGCAUUAGAAUGCUGCAAAAAAAU ............(....--)((((....))))((((((((((((((.......))).)))(((.((..(......)..)).)))))))))))........ ( -24.40) >consensus GAAAAGG___GAAAAUA__AGGACG_CGAGCUGGGGCGUUGCGUGUGAAAGGAGCAGCGCCUGUUGAAGGACGAACGGCAUCAGAAUGCUGCAGAAGAAA ..............................((((((((((((...........))))))))).............((((((....)))))))))...... (-20.67 = -21.08 + 0.42)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:42 2006