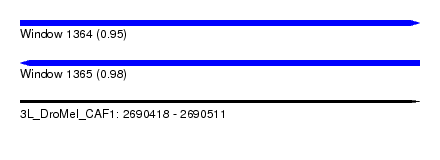
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,690,418 – 2,690,511 |
| Length | 93 |
| Max. P | 0.979011 |

| Location | 2,690,418 – 2,690,511 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 93.09 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -18.70 |
| Energy contribution | -18.46 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2690418 93 + 23771897 GCCAAACAAUGCGGCAAAUUUUGUUUGCCUACCGUGCACAAUGAGAAAGAGGUCCUUUCAUUCAUUACAGGGACC-A-AAAUGCAAUAUAUUCGC .........((((((((((...)))))))..((.((...((((((((((.....)))))..))))).)).))...-.-....))).......... ( -20.50) >DroSec_CAF1 51459 93 + 1 GCCAAACAAUGCGGCAAAUUUUGUUUGCCUACCGUGCACAAUGAGAAAGAGGUCCUUUCAUUCAUUACAAGGACC-A-AAAUGCAAUAUAUUCGC .........((((((((((...))))))).....................(((((((...........)))))))-.-....))).......... ( -20.10) >DroSim_CAF1 50323 93 + 1 GCCAAACAAUGCGGCAAAUUUUCUUUGCCUACCGUGCACAAUGAGAAAGAGGUCCUUUCAUUUAUUACAAGGACC-A-AAAUGCAAUAUAUUCGC .........(((((((((.....)))))).....................(((((((...........)))))))-.-....))).......... ( -19.60) >DroEre_CAF1 56300 94 + 1 GCCAAAUAAUGCGGCAAAUUUUGUUUGCCUACCGUGCACAAGGAAAAAGAGGUCCUUUCAUUCAUCACAGGGACCCA-AAAUGCACUGUAUUGGC ((((((((.((((((((((...)))))))..((........)).....(.(((((((...........)))))))).-....))).))).))))) ( -27.30) >DroYak_CAF1 52084 95 + 1 GCCAAAUAAUGCGGCAAAUUUUGUUUGCCUACCGUGCACAAGGAAAAAGAGGUCCUUUCACUCAUCACAAGGACCAAAAAAUGCAAUAUAUUCGC .....(((.((((((((((...)))))))..((........)).......(((((((...........))))))).......))).)))...... ( -21.80) >consensus GCCAAACAAUGCGGCAAAUUUUGUUUGCCUACCGUGCACAAUGAGAAAGAGGUCCUUUCAUUCAUUACAAGGACC_A_AAAUGCAAUAUAUUCGC .........(((((((((.....)))))).....................(((((((...........))))))).......))).......... (-18.70 = -18.46 + -0.24)

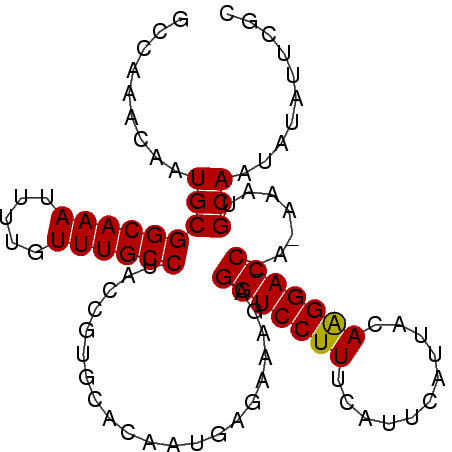
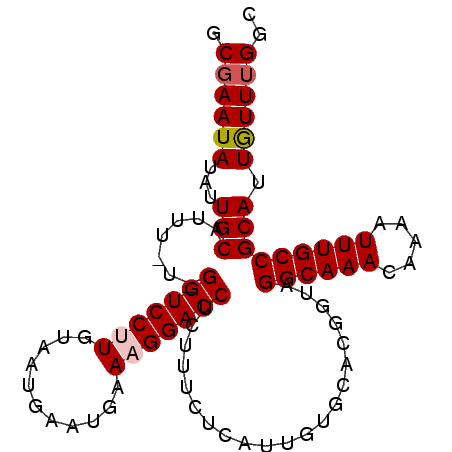
| Location | 2,690,418 – 2,690,511 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 93.09 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -22.72 |
| Energy contribution | -23.08 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979011 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2690418 93 - 23771897 GCGAAUAUAUUGCAUUU-U-GGUCCCUGUAAUGAAUGAAAGGACCUCUUUCUCAUUGUGCACGGUAGGCAAACAAAAUUUGCCGCAUUGUUUGGC .((((((...(((....-.-...((.((((...(((((((((....)))).))))).)))).))..((((((.....))))))))).)))))).. ( -23.90) >DroSec_CAF1 51459 93 - 1 GCGAAUAUAUUGCAUUU-U-GGUCCUUGUAAUGAAUGAAAGGACCUCUUUCUCAUUGUGCACGGUAGGCAAACAAAAUUUGCCGCAUUGUUUGGC .((((((...(((....-.-(((((((...........))))))).....................((((((.....))))))))).)))))).. ( -24.90) >DroSim_CAF1 50323 93 - 1 GCGAAUAUAUUGCAUUU-U-GGUCCUUGUAAUAAAUGAAAGGACCUCUUUCUCAUUGUGCACGGUAGGCAAAGAAAAUUUGCCGCAUUGUUUGGC .((((((...(((....-.-...((.((((...(((((((((....)))).))))).)))).))..((((((.....))))))))).)))))).. ( -26.00) >DroEre_CAF1 56300 94 - 1 GCCAAUACAGUGCAUUU-UGGGUCCCUGUGAUGAAUGAAAGGACCUCUUUUUCCUUGUGCACGGUAGGCAAACAAAAUUUGCCGCAUUAUUUGGC (((((...(((((....-.......((((((..(..((((((....))))))..)..).)))))..((((((.....)))))))))))..))))) ( -28.40) >DroYak_CAF1 52084 95 - 1 GCGAAUAUAUUGCAUUUUUUGGUCCUUGUGAUGAGUGAAAGGACCUCUUUUUCCUUGUGCACGGUAGGCAAACAAAAUUUGCCGCAUUAUUUGGC .((((((...(((............((((((..((..(((((....)))))..))..).)))))..((((((.....))))))))).)))))).. ( -26.20) >consensus GCGAAUAUAUUGCAUUU_U_GGUCCUUGUAAUGAAUGAAAGGACCUCUUUCUCAUUGUGCACGGUAGGCAAACAAAAUUUGCCGCAUUGUUUGGC .((((((...(((.......(((((((...........))))))).....................((((((.....))))))))).)))))).. (-22.72 = -23.08 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:38 2006