
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,686,838 – 2,686,942 |
| Length | 104 |
| Max. P | 0.967922 |

| Location | 2,686,838 – 2,686,942 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -35.77 |
| Consensus MFE | -19.63 |
| Energy contribution | -22.07 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.55 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967922 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
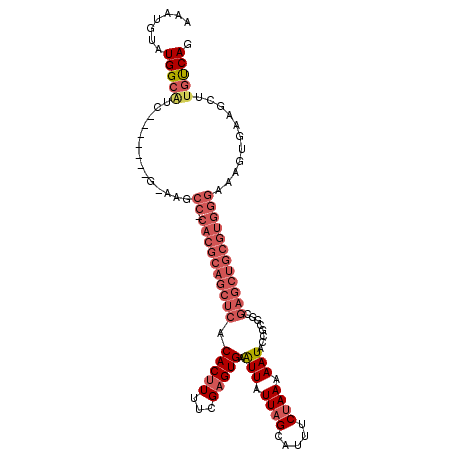
>3L_DroMel_CAF1 2686838 104 + 23771897 CUGACAAGCUUCACUUUCCCACGCAGCUCGCAGCGAUAUUUUUAGAAAUGCUAAUAAUUCACUCGAAAAGUGUGAGCUGCGUG-GGCUCCC-------GAUGCCAUACAUUU .......((.((.....(((((((((((((((.(((.(((.((((.....)))).)))....))).....)))))))))))))-)).....-------)).))......... ( -35.70) >DroSec_CAF1 47907 103 + 1 CUGACAAGCUUCACUUUCCCACGCAGCUCGCAGCGGUAUUUUUAGAAAUGCUAAUAAUUCACUCGAAAAGUGUGAGCUGCGUG-GGCUC-C-------GAUGCCAUACAUUU .......((.((.....(((((((((((((((..(((((((....))))))).....(((....)))...)))))))))))))-))...-.-------)).))......... ( -37.20) >DroSim_CAF1 46514 103 + 1 CUGACAAGCUUCACUUUCCCACGCAGCUCGCAGCGGUAUUUUUAGAAAUGCUAAUAAUUCACUCGAAAAGUGUGAGCUGCGUG-GGCUC-C-------GAUGCCAUACAUUU .......((.((.....(((((((((((((((..(((((((....))))))).....(((....)))...)))))))))))))-))...-.-------)).))......... ( -37.20) >DroEre_CAF1 52729 110 + 1 CUGACAAGCUUCGCUUUCCCACGCAGCUCGUCGCCGUAUUUUUAGAAAUGCUAAUAAUUCACUCGAAAAGUGUGAGCUGCGUG-GGCGU-AUGUAUGGGCUGCCAUACAUUU ...((((((...)))).(((((((((((((.(((.((((((....))))))......(((....)))..))))))))))))))-)).))-((((((((....)))))))).. ( -41.90) >DroYak_CAF1 48372 103 + 1 CUGACAAGCUUCACUUUCCCACGCAGCUCGCCGCGGUAUUUUUAGAAAUGCUAAUAAUUCACUCGAAAAGUGUGAGCUGCGUG-GGCGU-C-------GCAGCCACACAUUU .......(((.(((...((((((((((((((.(((((((((....))))))).....(((....)))..))))))))))))))-)).))-.-------).)))......... ( -37.00) >DroAna_CAF1 46172 90 + 1 CUGGCAGGCUUCGU-----------------CUAGAUAUUUUUGGAAAAGCUAAUAACUCACUCGAAAUGUGUGGGCUUCCUUGGGGGU-CCUU----CUCCCCACACAUUU ......(((((..(-----------------(((((....)))))).))))).............(((((((((((((((....)))).-....----...))))))))))) ( -25.60) >consensus CUGACAAGCUUCACUUUCCCACGCAGCUCGCAGCGGUAUUUUUAGAAAUGCUAAUAAUUCACUCGAAAAGUGUGAGCUGCGUG_GGCGC_C_______GAUGCCAUACAUUU ...................((((((((((((...(((((((....))))))).....(((....)))....)))))))))))).(((..............)))........ (-19.63 = -22.07 + 2.45)



| Location | 2,686,838 – 2,686,942 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.90 |
| Mean single sequence MFE | -35.03 |
| Consensus MFE | -20.26 |
| Energy contribution | -22.54 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2686838 104 - 23771897 AAAUGUAUGGCAUC-------GGGAGCC-CACGCAGCUCACACUUUUCGAGUGAAUUAUUAGCAUUUCUAAAAAUAUCGCUGCGAGCUGCGUGGGAAAGUGAAGCUUGUCAG ...((...(((.((-------(....((-((((((((((.(((((...)))))......((((...............)))).))))))))))))....))).)))...)). ( -38.26) >DroSec_CAF1 47907 103 - 1 AAAUGUAUGGCAUC-------G-GAGCC-CACGCAGCUCACACUUUUCGAGUGAAUUAUUAGCAUUUCUAAAAAUACCGCUGCGAGCUGCGUGGGAAAGUGAAGCUUGUCAG ...((...(((.((-------(-...((-((((((((((.(((((...)))))......((((...............)))).))))))))))))....))).)))...)). ( -37.86) >DroSim_CAF1 46514 103 - 1 AAAUGUAUGGCAUC-------G-GAGCC-CACGCAGCUCACACUUUUCGAGUGAAUUAUUAGCAUUUCUAAAAAUACCGCUGCGAGCUGCGUGGGAAAGUGAAGCUUGUCAG ...((...(((.((-------(-...((-((((((((((.(((((...)))))......((((...............)))).))))))))))))....))).)))...)). ( -37.86) >DroEre_CAF1 52729 110 - 1 AAAUGUAUGGCAGCCCAUACAU-ACGCC-CACGCAGCUCACACUUUUCGAGUGAAUUAUUAGCAUUUCUAAAAAUACGGCGACGAGCUGCGUGGGAAAGCGAAGCUUGUCAG ..((((((((....))))))))-...((-((((((((((.(((((...)))))........................(....))))))))))))).((((...))))..... ( -40.90) >DroYak_CAF1 48372 103 - 1 AAAUGUGUGGCUGC-------G-ACGCC-CACGCAGCUCACACUUUUCGAGUGAAUUAUUAGCAUUUCUAAAAAUACCGCGGCGAGCUGCGUGGGAAAGUGAAGCUUGUCAG .......((((.((-------.-((.((-((((((((((.(((((...))))).(((.((((.....)))).)))........))))))))))))...))...))..)))). ( -37.50) >DroAna_CAF1 46172 90 - 1 AAAUGUGUGGGGAG----AAGG-ACCCCCAAGGAAGCCCACACAUUUCGAGUGAGUUAUUAGCUUUUCCAAAAAUAUCUAG-----------------ACGAAGCCUGCCAG ((((((((((((..----....-..))))..((....))))))))))..............(((((((............)-----------------).)))))....... ( -17.80) >consensus AAAUGUAUGGCAUC_______G_AAGCC_CACGCAGCUCACACUUUUCGAGUGAAUUAUUAGCAUUUCUAAAAAUACCGCGGCGAGCUGCGUGGGAAAGUGAAGCUUGUCAG .......(((((..............((.((((((((((.(((((...))))).(((.((((.....)))).)))........))))))))))))...........))))). (-20.26 = -22.54 + 2.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:35 2006