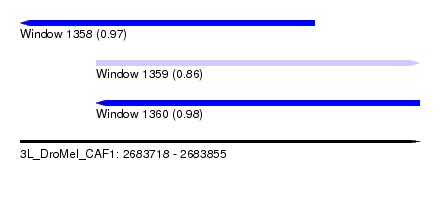
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 2,683,718 – 2,683,855 |
| Length | 137 |
| Max. P | 0.976292 |

| Location | 2,683,718 – 2,683,819 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 85.05 |
| Mean single sequence MFE | -26.34 |
| Consensus MFE | -18.08 |
| Energy contribution | -18.72 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2683718 101 - 23771897 GCUAAUUGGCAGCAUUUGUUUGCUCGACCAGUGCUGCCAAGCAAUGUCACGAAGCAGAUAUGUUUCUAAAUUCAUAAAUUGUACCAUACAAUUUAAUAGCA ((((.(((((((((((.(((.....))).)))))))))))..........((((((....))))))........(((((((((...))))))))).)))). ( -33.60) >DroSec_CAF1 44831 101 - 1 GCUAAUUGGCAGGAUUUGUUUGCUCGACCAGUGCUGCCAAGCAAUGUCACAAAGCAGAUCUGUUUCUAAAUUCAUAAAUUGUACCAUACAAUUCAAUACCA (((....))).((((((((((....(((...((((....))))..)))...))))))))))...............(((((((...)))))))........ ( -22.20) >DroSim_CAF1 43404 101 - 1 GCUAAUUGGCAGCAUUUGUUUGCUCGACCAGUGCUGCCAAGCAAUGUCACAAAGCAGAUCUGAUUCUAAAUUCAUAAAUUGUACCAUACAAUUCAAUACCA (((...((((((((((.(((.....))).))))))))))))).............(((......))).........(((((((...)))))))........ ( -23.30) >DroEre_CAF1 49682 101 - 1 GCAAAUUGGCAGCACAUGUUUGCUCGACCAGUGCUGCCAAGCUCAGCUACAAAGCAGAAUUGCUGCCUAAUUCAUAAAUUGUACCAUAAAAUCGCAAAGCA ((...((((((((((.((.((....)).))))))))))))((...(((....))).((((((.....))))))....................))...)). ( -28.10) >DroYak_CAF1 45162 101 - 1 GCAAAUUGGCAUCACUUGUUUGCCCGACCAGUGCUGCCAAGCUAGGCUACAAAGCAGAUCUGUUUCCUAACUCAAAAUUUUAACCAUAAAAUUGCAAAGCA ((...((((((.((((.(((.....))).)))).))))))((((((..(((.........)))..))))......(((((((....)))))))))...)). ( -24.50) >consensus GCUAAUUGGCAGCAUUUGUUUGCUCGACCAGUGCUGCCAAGCAAUGUCACAAAGCAGAUCUGUUUCUAAAUUCAUAAAUUGUACCAUACAAUUCAAUAGCA ((...(((((((((((.(((.....))).))))))))))))).........(((((....))))).................................... (-18.08 = -18.72 + 0.64)

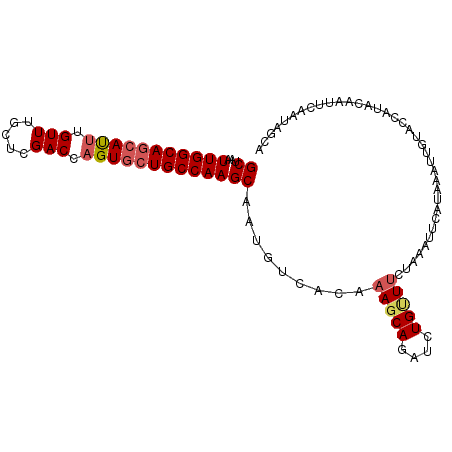
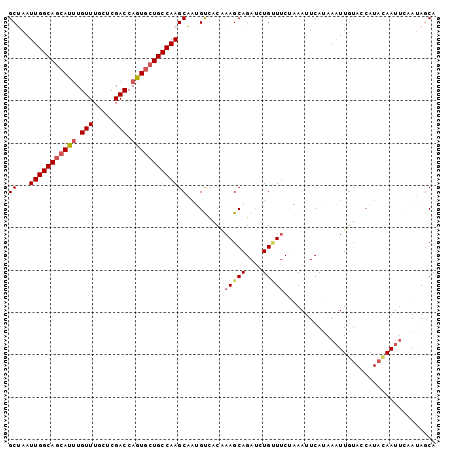
| Location | 2,683,744 – 2,683,855 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -17.38 |
| Energy contribution | -19.42 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 2683744 111 + 23771897 AUGAAUUUAGAAACAUAUCUGCUUCGUGACAUUGCUUGGCAGCACUGGUCGAGCAAACAAAUGCUGCCAAUUAGCAAGGACAUUUUUCAAAUGUUACUGUAAAAGAAAGAA ..(((..((((......)))).)))(((((((((((((((((((...((.......))...))))))))...)))..(((.....))).)))))))).............. ( -25.60) >DroSec_CAF1 44857 111 + 1 AUGAAUUUAGAAACAGAUCUGCUUUGUGACAUUGCUUGGCAGCACUGGUCGAGCAAACAAAUCCUGCCAAUUAGCAAGGACAUUUUUCAAAUGUUACUGUAGAAGAAAGAA .................(((((...(((((((((((((((.......))))))).......((((((......)).)))).........)))))))).)))))........ ( -28.80) >DroSim_CAF1 43430 111 + 1 AUGAAUUUAGAAUCAGAUCUGCUUUGUGACAUUGCUUGGCAGCACUGGUCGAGCAAACAAAUGCUGCCAAUUAGCAAGGACUUUAUUCAAAUGUUACUGUAGAAUAAAGAA .................(((((...(((((((((((((((((((...((.......))...))))))))...)))..(((.....))).)))))))).)))))........ ( -29.60) >DroEre_CAF1 49708 101 + 1 AUGAAUUAGGCAGCAAUUCUGCUUUGUAGCUGAGCUUGGCAGCACUGGUCGAGCAAACAUGUGCUGCCAAUUUGCAAGGAAAUCAACCAAAUCUUGUUGUA---------- ..(((((.(((((((..((.(((....))).))(((((((.......))))))).......))))))))))))((((((............))))))....---------- ( -30.00) >DroYak_CAF1 45188 109 + 1 UUGAGUUAGGAAACAGAUCUGCUUUGUAGCCUAGCUUGGCAGCACUGGUCGGGCAAACAAGUGAUGCCAAUUUGCAAGGAAAUCAAUCAAAUCUUGCUAAAGGACA--UAA ..((((((((..(((((.....)))))..))))))))((((.((((.((.......)).)))).)))).....((((((............)))))).........--... ( -29.30) >consensus AUGAAUUUAGAAACAGAUCUGCUUUGUGACAUUGCUUGGCAGCACUGGUCGAGCAAACAAAUGCUGCCAAUUAGCAAGGACAUUAUUCAAAUGUUACUGUAGAAGAAAGAA .................(((((...(((((((((((((((((((...((.......))...)))))))))...))..(((.....))).)))))))).)))))........ (-17.38 = -19.42 + 2.04)

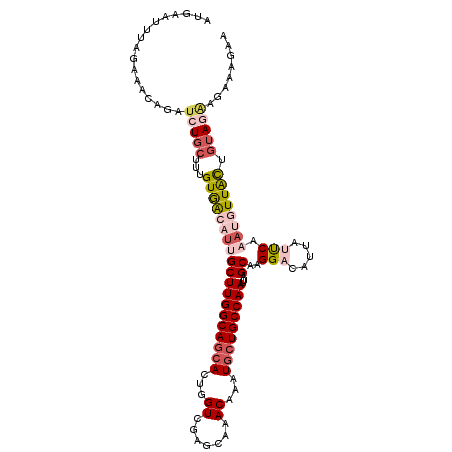
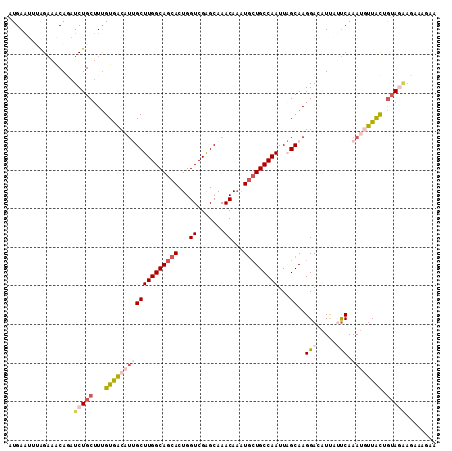
| Location | 2,683,744 – 2,683,855 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.28 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
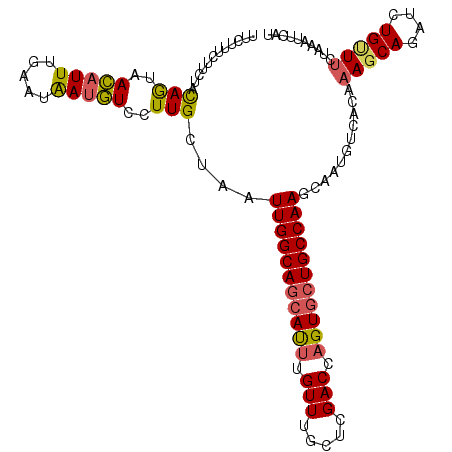
>3L_DroMel_CAF1 2683744 111 - 23771897 UUCUUUCUUUUACAGUAACAUUUGAAAAAUGUCCUUGCUAAUUGGCAGCAUUUGUUUGCUCGACCAGUGCUGCCAAGCAAUGUCACGAAGCAGAUAUGUUUCUAAAUUCAU ......................((((...((.(.(((((...((((((((((.(((.....))).))))))))))))))).).)).((((((....))))))....)))). ( -30.90) >DroSec_CAF1 44857 111 - 1 UUCUUUCUUCUACAGUAACAUUUGAAAAAUGUCCUUGCUAAUUGGCAGGAUUUGUUUGCUCGACCAGUGCUGCCAAGCAAUGUCACAAAGCAGAUCUGUUUCUAAAUUCAU ...........((((....(((((.....((.(.(((((...((((((.(((.(((.....))).))).))))))))))).).)).....)))))))))............ ( -24.00) >DroSim_CAF1 43430 111 - 1 UUCUUUAUUCUACAGUAACAUUUGAAUAAAGUCCUUGCUAAUUGGCAGCAUUUGUUUGCUCGACCAGUGCUGCCAAGCAAUGUCACAAAGCAGAUCUGAUUCUAAAUUCAU ..((((((((.............))))))))...(((((...((((((((((.(((.....))).)))))))))))))))...........(((......)))........ ( -28.12) >DroEre_CAF1 49708 101 - 1 ----------UACAACAAGAUUUGGUUGAUUUCCUUGCAAAUUGGCAGCACAUGUUUGCUCGACCAGUGCUGCCAAGCUCAGCUACAAAGCAGAAUUGCUGCCUAAUUCAU ----------............(((((((.......((...((((((((((.((.((....)).)))))))))))))))))))))....((((.....))))......... ( -27.61) >DroYak_CAF1 45188 109 - 1 UUA--UGUCCUUUAGCAAGAUUUGAUUGAUUUCCUUGCAAAUUGGCAUCACUUGUUUGCCCGACCAGUGCUGCCAAGCUAGGCUACAAAGCAGAUCUGUUUCCUAACUCAA ...--.((.....((((.((((((........(((.((...((((((.((((.(((.....))).)))).)))))))).)))........)))))))))).....)).... ( -27.29) >consensus UUCUUUCUUCUACAGUAACAUUUGAAUAAUGUCCUUGCUAAUUGGCAGCAUUUGUUUGCUCGACCAGUGCUGCCAAGCAAUGUCACAAAGCAGAUCUGUUUCUAAAUUCAU ............(((..(((((.....)))))..)))....(((((((((((.(((.....))).)))))))))))...........(((((....))))).......... (-19.48 = -19.28 + -0.20)

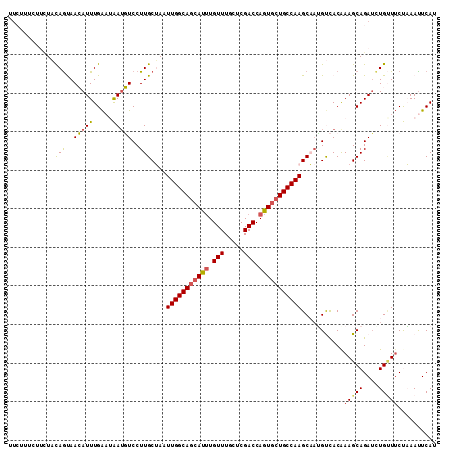
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:34 2006