
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 23,180,880 – 23,181,040 |
| Length | 160 |
| Max. P | 0.974156 |

| Location | 23,180,880 – 23,181,000 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -22.70 |
| Energy contribution | -22.30 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
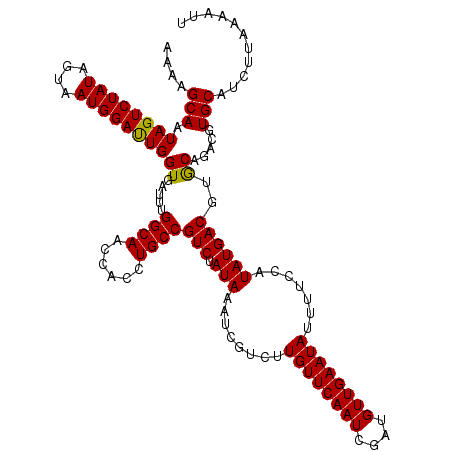
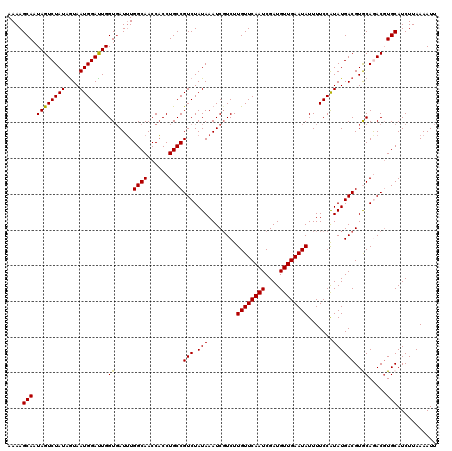
>3L_DroMel_CAF1 23180880 120 + 23771897 AAAAGCAAUAGUCUAUAAUAAUGGAUUGGUCAUAUGGCAACCUCCUGCCGUCUAUAAAUCCUCUUGUUCAAUCGAUGUUGAAUAUUUUCCGUAUGACGUACAAACCUGCAUCUUAAAAUU ....(((.((((((((....))))))))(((((((((((......))))...............((((((((....))))))))......))))))).........)))........... ( -26.80) >DroGri_CAF1 31676 120 + 1 AAAAGCAAUAGUCUAUAGUAAUGGAUUGGUGAUUUGGCAACCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCAUAUGACGUGCAGACGUGCAUCUUAAAAUU ....(((...((((...((.(((((..(..(((((((((......))))......)))))..).((((((((....))))))))...)))))...))....)))).)))........... ( -27.10) >DroWil_CAF1 25720 120 + 1 AAAAGCAAUAGUCUAUAGUAAUGGAUUGGUGAUUUGGCAACCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCAUAUGACGUGCAGACGUGCAUCUUAAAAUU ....(((...((((...((.(((((..(..(((((((((......))))......)))))..).((((((((....))))))))...)))))...))....)))).)))........... ( -27.10) >DroMoj_CAF1 13757 120 + 1 AAAAGCAAUAGUCUAUAGUAAUGGAUUGGUGAUUUGGCAACCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCAUAUGACGUGCAGACGUGCAUCUUAAAAUU ....(((...((((...((.(((((..(..(((((((((......))))......)))))..).((((((((....))))))))...)))))...))....)))).)))........... ( -27.10) >DroPer_CAF1 52661 120 + 1 AAAAGCAAUAGUCUAUAGUAAUGGACUGGUGAUUUGGCAACCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCGUAUGACGUACAAACGUGCAUCUUAAAAUU .....((.((((((((....)))))))).))....((((......))))(((.(((........((((((((....)))))))).......))))))((((....))))........... ( -26.46) >consensus AAAAGCAAUAGUCUAUAGUAAUGGAUUGGUGAUUUGGCAACCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCAUAUGACGUGCAGACGUGCAUCUUAAAAUU ....(((.((((((((....))))))))((.....((((......))))(((.(((........((((((((....)))))))).......))))))..)).....)))........... (-22.70 = -22.30 + -0.40)

| Location | 23,180,920 – 23,181,040 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -29.79 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.92 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974156 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
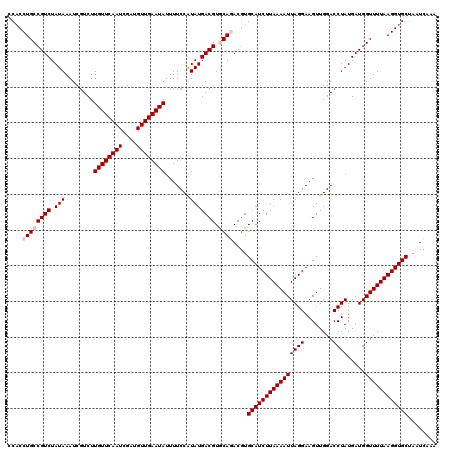
>3L_DroMel_CAF1 23180920 120 + 23771897 CCUCCUGCCGUCUAUAAAUCCUCUUGUUCAAUCGAUGUUGAAUAUUUUCCGUAUGACGUACAAACCUGCAUCUUAAAAUUAGGAAGUUGGACCUAUGAUGGUUUUAAGGUGCUACUCAAA ........((((.(((........((((((((....)))))))).......))))))).........((((((((((((((((........)))).....))))))))))))........ ( -25.26) >DroGri_CAF1 31716 120 + 1 CCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCAUAUGACGUGCAGACGUGCAUCUUAAAAUUAGGAAGUUGGACCUAUGAUGGUUUUAAGGUGCUAUUCAAA ....((((((((.(((........((((((((....)))))))).......))))))).))))....((((((((((((((((........)))).....))))))))))))........ ( -31.76) >DroWil_CAF1 25760 120 + 1 CCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCAUAUGACGUGCAGACGUGCAUCUUAAAAUUAGGAAGUUGGACCUAUGAUGGUUUUAAGGUGCUACUCAAA ....((((((((.(((........((((((((....)))))))).......))))))).))))..((((((((((((((((((........)))).....)))))))))))).))..... ( -32.56) >DroMoj_CAF1 13797 120 + 1 CCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCAUAUGACGUGCAGACGUGCAUCUUAAAAUUAGGAAGUUGGACCUAUGAUGGUUUUAAGGUGCUAAUCAAA ....((((((((.(((........((((((((....)))))))).......))))))).))))....((((((((((((((((........)))).....))))))))))))........ ( -31.76) >DroPer_CAF1 52701 120 + 1 CCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCGUAUGACGUACAAACGUGCAUCUUAAAAUUAGGAAGUUGGACCUAUGAUGGUUUUAAGGUGCUAAUCUAA .(((((((((((.(((....((((((((((((....)))))))).(((((.((.((.((((....)))).)).))......)))))..)))).)))))))))....)))))......... ( -27.60) >consensus CCACCUGCCGUCUAUAAAUCGUCUUGUUCAAUCGAUGUUGAAUAUUUUCCAUAUGACGUGCAGACGUGCAUCUUAAAAUUAGGAAGUUGGACCUAUGAUGGUUUUAAGGUGCUAAUCAAA ....((((((((.(((........((((((((....)))))))).......))))))).))))....((((((((((((((((........)))).....))))))))))))........ (-27.12 = -27.92 + 0.80)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:58 2006