
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,865,400 – 22,865,514 |
| Length | 114 |
| Max. P | 0.713673 |

| Location | 22,865,400 – 22,865,493 |
|---|---|
| Length | 93 |
| Sequences | 3 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.79 |
| Mean single sequence MFE | -26.23 |
| Consensus MFE | -18.14 |
| Energy contribution | -20.25 |
| Covariance contribution | 2.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
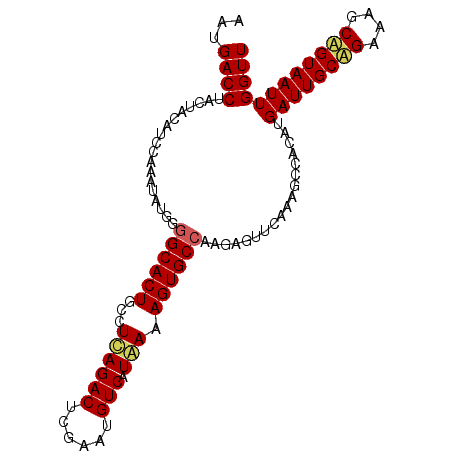
>3L_DroMel_CAF1 22865400 93 + 23771897 --------------------AUCGAUUGUGGUCCCCUCAAUGACCUACUACAACCAAAUAUGCGGCACUGCUUCAGACUCGAAUGUCAUAAAAGUGCAAAGAGUUCAAA-----UUGA --------------------.((((((((((((........)))).))................(((((......(((......))).....)))))..........))-----)))) ( -17.60) >DroSec_CAF1 53906 118 + 1 AACAACGUGUUGCUAUUCAAAUCGAUUUCGGUCCCCUCAAUGACCUACUACAUCCAAAUAUGGGGCACUGCCUCAGACUCGAAUGUCAUGAAAGUGCCAAGAGUUCAAAGCCACAUGA .....(((((.(((((((...........((((........))))........(((....)))((((((...((((((......))).))).))))))..))))....))).))))). ( -31.70) >DroSim_CAF1 57813 118 + 1 AACAACGUGUUGCUAUUCAAAUCGAUUUCGGUCCCCUCAAUGACCUACUACAUCCAAAUAUGGGGCACUGCCUUAGACUCGAAUGUCAUAAAAGUGCCAAGAGUUCAAAGCCACAUGA .....(((((.(((((((...........((((........))))........(((....)))((((((...((((((......))).))).))))))..))))....))).))))). ( -29.40) >consensus AACAACGUGUUGCUAUUCAAAUCGAUUUCGGUCCCCUCAAUGACCUACUACAUCCAAAUAUGGGGCACUGCCUCAGACUCGAAUGUCAUAAAAGUGCCAAGAGUUCAAAGCCACAUGA .....(((((.(((.......((......((((........))))..................((((((...((((((......))).))).))))))..))......))).))))). (-18.14 = -20.25 + 2.11)



| Location | 22,865,418 – 22,865,514 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 89.22 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -20.27 |
| Energy contribution | -20.83 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22865418 96 + 23771897 AAUGACCUACUACAACCAAAUAUGCGGCACUGCUUCAGACUCGAAUGUCAUAAAAGUGCAAAGAGUUCAAA-----UUGAUUGCUGG-AGCAGUAAUUGGUU ...((((........((........)).((((((((((((((...((.(((....)))))..))))((...-----..))...))))-))))))....)))) ( -23.10) >DroSec_CAF1 53944 102 + 1 AAUGACCUACUACAUCCAAAUAUGGGGCACUGCCUCAGACUCGAAUGUCAUGAAAGUGCCAAGAGUUCAAAGCCACAUGAUUGCAGAAAGCUGUAAUUGGUU ..(((.((.((....(((....)))((((((...((((((......))).))).)))))).)))).))).(((((....(((((((....)))))))))))) ( -29.10) >DroSim_CAF1 57851 102 + 1 AAUGACCUACUACAUCCAAAUAUGGGGCACUGCCUUAGACUCGAAUGUCAUAAAAGUGCCAAGAGUUCAAAGCCACAUGAUUGCAGUAAGCAGUAAUUGGUU ..(((.((.((....(((....)))((((((...((((((......))).))).)))))).)))).))).(((((....(((((.....)))))...))))) ( -26.40) >consensus AAUGACCUACUACAUCCAAAUAUGGGGCACUGCCUCAGACUCGAAUGUCAUAAAAGUGCCAAGAGUUCAAAGCCACAUGAUUGCAGAAAGCAGUAAUUGGUU ...((((..................((((((...((((((......))).))).))))))..................((((((((....)))))))))))) (-20.27 = -20.83 + 0.56)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:43 2006