
| Sequence ID | 3L_DroMel_CAF1 |
|---|---|
| Location | 22,781,532 – 22,781,661 |
| Length | 129 |
| Max. P | 0.975952 |

| Location | 22,781,532 – 22,781,622 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -24.02 |
| Energy contribution | -24.18 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975952 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22781532 90 + 23771897 GGUGGCCAUCAGCUGCA---------AAAA-AUCAUUUCAC-A----CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCA ((((((..((.......---------....-.........(-(----.(((((.....))))).)).(((((((...)))))))))..)))((....))..))). ( -31.10) >DroSec_CAF1 44215 91 + 1 GGUGGCCAUCAGCUGCA---------AAAAUAUAAUCUCAU-A----CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCA ((((((..((.......---------...........(((.-.----.(((((.....))))).)))(((((((...)))))))))..)))((....))..))). ( -30.60) >DroSim_CAF1 46308 91 + 1 GGUGGCCAUCAGCUGCA---------AAAAAAAAAUCUCAU-A----CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCA ((((((..((.......---------...........(((.-.----.(((((.....))))).)))(((((((...)))))))))..)))((....))..))). ( -30.60) >DroEre_CAF1 43449 92 + 1 UUCACACAUUAGUUAAA---------AAAAAAGUCUUUCAC-A---CCUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCA .................---------......(.(((((..-.---..(((((.....)))))....(((((((...)))))))))))).).(((......))). ( -25.90) >DroYak_CAF1 46725 104 + 1 -UGGCAAACCAGCUAAACUACAAUAUAAAAAAAUCUUUUAAGACUGACUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCA -.(((...........................................(((((.....)))))((..(((((((...)))))))..))))).(((......))). ( -27.30) >consensus GGUGGCCAUCAGCUGCA_________AAAAAAAAAUUUCAC_A____CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCA .(((((.....)))))...................((((.........(((((.....)))))....(((((((...)))))))))))....(((......))). (-24.02 = -24.18 + 0.16)



| Location | 22,781,553 – 22,781,661 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -36.92 |
| Consensus MFE | -30.47 |
| Energy contribution | -30.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22781553 108 + 23771897 -AUCAUUUCAC-A-----CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAGUUGAAAUAUAUGUG-UAUAAACUA -...((((((.-(-----((.((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))))))))))).......-......... ( -35.60) >DroVir_CAF1 57461 113 + 1 UAAGAGU--GA-AGAUCACUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAUGGUUAGUUUAGGCGGCAAAAAUGC .......--..-...(((.(((((.....))))).)))(((((((...)))))))....((((..((..((((((((((.......)))..)))))))..))..))))........ ( -40.10) >DroGri_CAF1 61672 113 + 1 -AAGAUUUCAA-AGAUC-UUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAUGGUUGAUUUUGCUCAUCGGAAACC -..(((....(-(((((-..(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))).....))))))....)))(....). ( -37.10) >DroSim_CAF1 46330 108 + 1 -AAAAUCUCAU-A-----CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAGAUGUGAGGUUGACG-UGUAAACUA -..((((((((-(-----.((((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....)))))))).)))))))))....-......... ( -42.30) >DroEre_CAF1 43471 109 + 1 -AGUCUUUCAC-A---C-CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAUGUACAAUUCAGUCG-UGUAAACUA -........((-(---(-..(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....))))))).(.((......))))-)))...... ( -32.50) >DroYak_CAF1 46755 113 + 1 -AAUCUUUUAAGACUGA-CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAUGCACAAUUUUUUCG-UGUAAACUA -................-..(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....)))))))(((((.........)-))))..... ( -33.90) >consensus _AAGAUUUCAA_A___C_CUUCCCAUACCGGGAAUUGAACCCGGGCCUCCCGGGUGAGAGCCGGGUAUCCUAACCACUAGACAAUAUGGGAUGUAUAAUUUAGGCG_UGUAAACUA ....................(((((((((((...((..(((((((...)))))))..)).)))).....(((.....)))....)))))))......................... (-30.47 = -30.47 + -0.00)

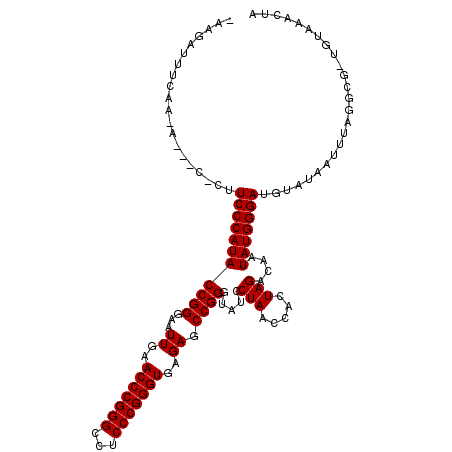

| Location | 22,781,553 – 22,781,661 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -36.84 |
| Consensus MFE | -30.93 |
| Energy contribution | -30.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3L_DroMel_CAF1 22781553 108 - 23771897 UAGUUUAUA-CACAUAUAUUUCAACUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAG-----U-GUGAAAUGAU- .........-......(((((((((((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))).))-----)-.)))))))..- ( -37.00) >DroVir_CAF1 57461 113 - 1 GCAUUUUUGCCGCCUAAACUAACCAUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAGUGAUCU-UC--ACUCUUA (((....))).((((...((((((((.............))))))))....(((((.......)))))))))(((((.......)))))..(((((.(((....-.)--))))))) ( -38.62) >DroGri_CAF1 61672 113 - 1 GGUUUCCGAUGAGCAAAAUCAACCAUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAA-GAUCU-UUGAAAUCUU- ((((((.(((.......))).....((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))..-.....-..))))))..- ( -36.50) >DroSim_CAF1 46330 108 - 1 UAGUUUACA-CGUCAACCUCACAUCUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAG-----U-AUGAGAUUUU- .........-.......((((.(..((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))..-----)-.)))).....- ( -36.50) >DroEre_CAF1 43471 109 - 1 UAGUUUACA-CGACUGAAUUGUACAUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAG-G---U-GUGAAAGACU- ...((((((-(.((......)).(.((((((((((......((....((.(((((((((((....)))))..))))))))....)))))))))))).)-)---)-))))).....- ( -37.80) >DroYak_CAF1 46755 113 - 1 UAGUUUACA-CGAAAAAAUUGUGCAUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAG-UCAGUCUUAAAAGAUU- .......((-(((.....)))))..((((((((((......((....((.(((((((((((....)))))..))))))))....))))))))))))..-..(((((....)))))- ( -34.60) >consensus UAGUUUACA_CGACAAAAUUAAACAUCCCAUAUUGUCUAGUGGUUAGGAUACCCGGCUCUCACCCGGGAGGCCCGGGUUCAAUUCCCGGUAUGGGAAG_G___U_UUGAAAUAUU_ .........................((((((((((......((....((.(((((((((((....)))))..))))))))....)))))))))))).................... (-30.93 = -30.93 + 0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:23:21 2006